 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Itr_sc000505.1_g00006.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; lamiids; Solanales; Convolvulaceae; Ipomoeeae; Ipomoea
|
||||||||
| Family | AP2 | ||||||||
| Protein Properties | Length: 618aa MW: 66857.4 Da PI: 7.2206 | ||||||||
| Description | AP2 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | AP2 | 33.5 | 1.1e-10 | 225 | 281 | 1 | 55 |
AP2 1 sgykGVrwdkkrgrWvAeIrd.pseng..kr.krfslgkfgtaeeAakaaiaarkkleg 55
s y+GV++++++gr++A+++d + ++ k + g ++ +++Aa+a++ a++k++g
Itr_sc000505.1_g00006.1 225 SIYRGVTRHRWTGRYEAHLWDnSCR-RegQSrKGRQ-GGYDKEDKAARAYDLAALKYWG 281
57*******************5444.2455535555.7799999*************98 PP
| |||||||
| 2 | AP2 | 46.9 | 7e-15 | 326 | 375 | 3 | 55 |
AP2 3 ykGVrwdkkrgrWvAeIrdpsengkrkrfslgkfgtaeeAakaaiaarkkleg 55
y+GV+++++ grW A+I + +k +lg+f t+eeAa+a++ a+ k++g
Itr_sc000505.1_g00006.1 326 YRGVTRHHQHGRWQARIGRVAG---NKDLYLGTFSTEEEAAEAYDIAAIKFRG 375
9***************988532...5************************998 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF00847 | 3.2E-8 | 225 | 281 | IPR001471 | AP2/ERF domain |
| SuperFamily | SSF54171 | 3.27E-14 | 225 | 291 | IPR016177 | DNA-binding domain |
| CDD | cd00018 | 2.33E-18 | 225 | 291 | No hit | No description |
| SMART | SM00380 | 2.6E-22 | 226 | 295 | IPR001471 | AP2/ERF domain |
| PROSITE profile | PS51032 | 17.201 | 226 | 289 | IPR001471 | AP2/ERF domain |
| Gene3D | G3DSA:3.30.730.10 | 1.9E-11 | 226 | 290 | IPR001471 | AP2/ERF domain |
| PRINTS | PR00367 | 3.0E-6 | 227 | 238 | IPR001471 | AP2/ERF domain |
| CDD | cd00018 | 1.07E-23 | 324 | 385 | No hit | No description |
| SuperFamily | SSF54171 | 6.34E-18 | 324 | 385 | IPR016177 | DNA-binding domain |
| SMART | SM00380 | 8.6E-33 | 325 | 389 | IPR001471 | AP2/ERF domain |
| PROSITE profile | PS51032 | 19.375 | 325 | 383 | IPR001471 | AP2/ERF domain |
| Gene3D | G3DSA:3.30.730.10 | 3.6E-18 | 325 | 384 | IPR001471 | AP2/ERF domain |
| Pfam | PF00847 | 1.7E-9 | 326 | 375 | IPR001471 | AP2/ERF domain |
| PRINTS | PR00367 | 3.0E-6 | 365 | 385 | IPR001471 | AP2/ERF domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0000723 | Biological Process | telomere maintenance | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0007389 | Biological Process | pattern specification process | ||||
| GO:0010449 | Biological Process | root meristem growth | ||||
| GO:0019827 | Biological Process | stem cell population maintenance | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 618 aa Download sequence Send to blast |
MGSMNSNNWL SFPLSPSQAA AQSHHFSVGL VNDSLDTSFQ NPEVMISPEP APVPPGANVG 60 GPQTVKRTER KTSLGARLGH YWPFAQHKWN LISTQGSNEV PKVADFLGVG KVDCNQPDLV 120 SYNDIQANEP DYLFSSNAGG LLPVQTGGVT VAAGAGGGFE LQENNACNMQ SLTLSMGTSG 180 KGSATTHASP SGNATASGSA ENSNTSIVEA ATPRRALDTF GQRTSIYRGV TRHRWTGRYE 240 AHLWDNSCRR EGQSRKGRQG GYDKEDKAAR AYDLAALKYW GTSTTTNFPI SNYEKELEEM 300 KHMTRQEFVA AIRRKSSGFS RGASMYRGVT RHHQHGRWQA RIGRVAGNKD LYLGTFSTEE 360 EAAEAYDIAA IKFRGLNAVT NFDMSRYDVK AILESNTLPI GGGAAKRLKE AQALESSRKR 420 DQEMLALNSS FHYSATTGPI HAAAAAAAYP LMQPFDAQPL LTLQNQDFSH HYAPQESPYT 480 YIQTHLQLHH QQQQHSPSYL NPSPQTSQYY NNNNNNNNNN PYLQSGNPGL LHGLMNMGGS 540 SSSVMDNNGG TSSGSYSGGG YLGNSTLGMA TNSNSGGGGD QELAVVKVDY DMPSGTYAGW 600 SNDSVQGSNP NIFSMWND |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probably acts as a transcriptional activator. Binds to the GCC-box pathogenesis-related promoter element. May be involved in the regulation of gene expression by stress factors and by components of stress signal transduction pathways (By similarity). Master regulator of basal/root fate. Essential for root quiescent center (QC) and columella specification, stem cell activity, as well as for establishment of the stem cell niche during embryogenesis. Modulates the root polar auxin transport by regulating the distribution of PIN genes. Essential role in respecifying pattern and polarity in damaged roots. Direct target of the transcriptional corepressor TPL. Expression levels and patterns regulated post-transcriptionally by root meristem growth factors (RGFs). {ECO:0000250, ECO:0000269|PubMed:15454085, ECO:0000269|PubMed:15635403, ECO:0000269|PubMed:16424342, ECO:0000269|PubMed:20190735}. | |||||
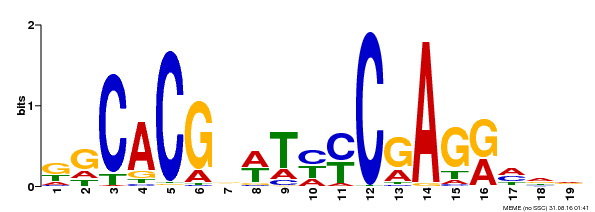
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00368 | DAP | Transfer from AT3G20840 | Download |
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: By auxin accumulation. {ECO:0000269|PubMed:15454085}. | |||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_019163643.1 | 0.0 | PREDICTED: AP2-like ethylene-responsive transcription factor PLT1 | ||||
| Refseq | XP_019163644.1 | 0.0 | PREDICTED: AP2-like ethylene-responsive transcription factor PLT1 | ||||
| Swissprot | Q5YGP8 | 0.0 | PLET1_ARATH; AP2-like ethylene-responsive transcription factor PLT1 | ||||
| TrEMBL | A0A068VG92 | 0.0 | A0A068VG92_COFCA; Uncharacterized protein | ||||
| STRING | EOY32584 | 0.0 | (Theobroma cacao) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Asterids | OGEA2246 | 24 | 57 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G20840.1 | 0.0 | AP2 family protein | ||||



