 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Itr_sc001316.1_g00005.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; lamiids; Solanales; Convolvulaceae; Ipomoeeae; Ipomoea
|
||||||||
| Family | HD-ZIP | ||||||||
| Protein Properties | Length: 823aa MW: 88866.6 Da PI: 5.8947 | ||||||||
| Description | HD-ZIP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Homeobox | 67.1 | 2.4e-21 | 135 | 190 | 1 | 56 |
TT--SS--HHHHHHHHHHHHHSSS--HHHHHHHHHHCTS-HHHHHHHHHHHHHHHH CS
Homeobox 1 rrkRttftkeqleeLeelFeknrypsaeereeLAkklgLterqVkvWFqNrRakek 56
+++ +++t++q++eLe+lF+++++p++++r eL+k+l L+ rqVk+WFqNrR+++k
Itr_sc001316.1_g00005.1 135 KKRYHRHTPQQIQELEALFKECPHPDEKQRSELSKRLCLENRQVKFWFQNRRTQMK 190
688999***********************************************999 PP
| |||||||
| 2 | START | 184.1 | 7.7e-58 | 333 | 555 | 1 | 206 |
HHHHHHHHHHHHHHHHC-TT-EEEE....EXCCTTEEEEEEESSS......SCEEEEEEEECCSCHHHHHHHHHCCCGGCT-TT-S CS
START 1 elaeeaaqelvkkalaeepgWvkss....esengdevlqkfeeskv.....dsgealrasgvvdmvlallveellddkeqWdetla 77
ela++a++elvk a+ +ep+Wv+ + e n++e+l++f++ + + +ea+r++g+v+ ++ lve+l+d + +W e+++
Itr_sc001316.1_g00005.1 333 ELALAAMDELVKVAQTDEPLWVRGVeggrEVMNQEEYLRSFSPCIGlrpsgFVSEASRETGMVIINSLALVETLMDCG-KWGEMFP 417
5899*********************9999999*********99999********************************.******* PP
....EEEEEEEECTT......EEEEEEEEXXTTXX-SSX.EEEEEEEEEEE.TTS-EEEEEEEEE-TTS--.-TTSEE-EESSEEE CS
START 78 ....kaetlevissg......galqlmvaelqalsplvp.RdfvfvRyirqlgagdwvivdvSvdseqkppesssvvRaellpSgi 152
+ +t++vis g galqlm+aelq+lsplvp R++ f+R+++q+ +g+w++vdvS+d ++ + ++ R +lpSg+
Itr_sc001316.1_g00005.1 418 cliaRTSTTDVISGGmggtrnGALQLMYAELQVLSPLVPiRKVNFLRFCKQHAEGVWAVVDVSIDAIRQSSAAPNATR--RLPSGC 501
********************************************************************9998999999..****** PP
EEEEECTCEEEEEEEE-EE--SSXXHHHHHHHHHHHHHHHHHHHHHHTXXXXXX CS
START 153 liepksnghskvtwvehvdlkgrlphwllrslvksglaegaktwvatlqrqcek 206
++++++ng+skvtwveh++++++ +h+l+r l++ g+ +ga++w atlq+qce+
Itr_sc001316.1_g00005.1 502 VVQDMPNGYSKVTWVEHAEYDESEVHQLYRGLIGAGMGFGAQRWIATLQQQCEC 555
****************************************************96 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:1.10.10.60 | 5.4E-22 | 121 | 192 | IPR009057 | Homeodomain-like |
| SuperFamily | SSF46689 | 3.25E-20 | 122 | 192 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS50071 | 17.346 | 132 | 192 | IPR001356 | Homeobox domain |
| SMART | SM00389 | 1.9E-17 | 133 | 196 | IPR001356 | Homeobox domain |
| CDD | cd00086 | 2.78E-18 | 134 | 192 | No hit | No description |
| Pfam | PF00046 | 8.3E-19 | 135 | 190 | IPR001356 | Homeobox domain |
| PROSITE pattern | PS00027 | 0 | 167 | 190 | IPR017970 | Homeobox, conserved site |
| PROSITE profile | PS50848 | 39.06 | 324 | 558 | IPR002913 | START domain |
| SuperFamily | SSF55961 | 3.48E-30 | 326 | 555 | No hit | No description |
| CDD | cd08875 | 6.06E-114 | 328 | 554 | No hit | No description |
| SMART | SM00234 | 6.5E-43 | 333 | 555 | IPR002913 | START domain |
| Pfam | PF01852 | 3.1E-50 | 333 | 555 | IPR002913 | START domain |
| SuperFamily | SSF55961 | 1.1E-24 | 582 | 816 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009827 | Biological Process | plant-type cell wall modification | ||||
| GO:0042335 | Biological Process | cuticle development | ||||
| GO:0043481 | Biological Process | anthocyanin accumulation in tissues in response to UV light | ||||
| GO:0048765 | Biological Process | root hair cell differentiation | ||||
| GO:0008289 | Molecular Function | lipid binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 823 aa Download sequence Send to blast |
MNFGGFLDNS SSIGSGGEGD VVGGGARIVA DIPYNSSSDM ASGAIAQPSL VPHSLTTKSM 60 FNSPGLSLGL QTGMEGGQGE VARVPDSYEA NNSNTNNSVG RRSREEEPES RSGSDNFEGA 120 SGDDQDAADD KPPRKKRYHR HTPQQIQELE ALFKECPHPD EKQRSELSKR LCLENRQVKF 180 WFQNRRTQMK TQLERHENSI LRQENDKLRA ENMSIREAMR NPLCTNCGGP AMIGEISLEE 240 QHLRIENVRL KDELDRVCAL AGKFLGRPVS SLPNINSSLE LGVGGNGFGG LSVVPSTVPI 300 APPPPPDFAG ALPTRQGSGV GIERSLERSM YLELALAAMD ELVKVAQTDE PLWVRGVEGG 360 REVMNQEEYL RSFSPCIGLR PSGFVSEASR ETGMVIINSL ALVETLMDCG KWGEMFPCLI 420 ARTSTTDVIS GGMGGTRNGA LQLMYAELQV LSPLVPIRKV NFLRFCKQHA EGVWAVVDVS 480 IDAIRQSSAA PNATRRLPSG CVVQDMPNGY SKVTWVEHAE YDESEVHQLY RGLIGAGMGF 540 GAQRWIATLQ QQCECLAILM SSALPTRDHT AISPSGRRSM LKLAQRMTNN FCAGVCASSV 600 HQWNKLCAGN VDEDVRVMTR KSVDDPGEPS GIVLSAATSV WLPVSPQRLF DFLRDERLRS 660 EWDILSNGGP MHEMAHIAKG QDHGNCVSLL RASAMSSNQS SMLILQETCI NAAGSLVVYA 720 PVDIPAMHVV MNGGDSAYVA LLPSGFAIVP DGPGSRGPAS ATNGPCSNGG PYQRVSGSLL 780 TVAFQILVNS LPTAKLTVES VETVNNLISC TVQKIKAALH CES |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcription factor involved in the regulation of the tissue-specific accumulation of anthocyanins and in cellular organization of the primary root. {ECO:0000269|PubMed:10402424}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
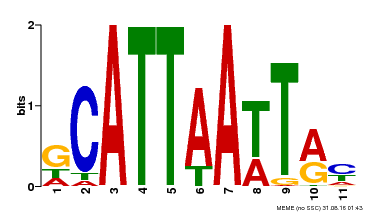
| Motif ID | Method | Source | Motif file |
| MP00421 | DAP | Transfer from AT4G00730 | Download |
| |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_019152252.1 | 0.0 | PREDICTED: homeobox-leucine zipper protein ANTHOCYANINLESS 2-like | ||||
| Swissprot | Q0WV12 | 0.0 | ANL2_ARATH; Homeobox-leucine zipper protein ANTHOCYANINLESS 2 | ||||
| TrEMBL | A0A2G3DE11 | 0.0 | A0A2G3DE11_CAPCH; Homeobox-leucine zipper protein ANTHOCYANINLESS 2 | ||||
| STRING | Solyc01g091630.2.1 | 0.0 | (Solanum lycopersicum) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Asterids | OGEA1626 | 24 | 65 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G61150.1 | 0.0 | homeodomain GLABROUS 1 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



