 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Itr_sc001419.1_g00007.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; lamiids; Solanales; Convolvulaceae; Ipomoeeae; Ipomoea
|
||||||||
| Family | bZIP | ||||||||
| Protein Properties | Length: 485aa MW: 52240.6 Da PI: 6.6617 | ||||||||
| Description | bZIP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | bZIP_1 | 31.5 | 4e-10 | 270 | 313 | 13 | 56 |
HHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHH CS
bZIP_1 13 NReAArrsRqRKkaeieeLeekvkeLeaeNkaLkkeleelkkev 56
NR +A rs +RK +i+eLe+kv++L++e ++L +l l+ +v
Itr_sc001419.1_g00007.1 270 NRQSAARSKERKMRYIAELERKVQTLQTEATSLSAQLTLLQAMV 313
*************************************9999887 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SMART | SM00338 | 4.7E-5 | 262 | 325 | IPR004827 | Basic-leucine zipper domain |
| CDD | cd14703 | 1.70E-22 | 267 | 311 | No hit | No description |
| SuperFamily | SSF57959 | 3.86E-9 | 267 | 311 | No hit | No description |
| PROSITE profile | PS50217 | 8.531 | 270 | 310 | IPR004827 | Basic-leucine zipper domain |
| Gene3D | G3DSA:1.20.5.170 | 2.2E-10 | 270 | 312 | No hit | No description |
| Pfam | PF00170 | 3.8E-8 | 270 | 313 | IPR004827 | Basic-leucine zipper domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 485 aa Download sequence Send to blast |
MDKDKSPALG GGLLPPSGRY SHFSQPGNGF SGKPEPSGSF NLPPLGPSAS ASESGHFGIG 60 VPSESNRFSQ DISQMPDNPP KKLGHRRAHS EIITLPDDIS FDSDLGVVGG LDGPSLSDET 120 EEDLFSMYLD MDKFNSSSAT SPLQVGGSSS SVATAPGSSQ APALVTPIAE NATSAVSEKP 180 RVRHQHSQSM DGSTTIKPEM LTSGTDEPSA AESKKAISAA KLAELALIDP KRAKRGGWGF 240 LIPGRTFPFR NTDFTVGASV LTPDLQIWAN RQSAARSKER KMRYIAELER KVQTLQTEAT 300 SLSAQLTLLQ AMVGAVLGRI CFLLSNPVRD TTGLTAENSE LKLRLQTMEQ QVHLQDALND 360 ALKEEIQHLK VLTGQGMANG GPMMMNFGPS FGAGQQFYPN NHAMHTLLTA HQLQQLQLHS 420 QKPQQPPNFQ QHQLHPFQLQ PQQQPPPLQP QEQHLHQAGD PKLRSSLSAS ALTEHASDSC 480 TSMKD |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Putative transcription factor with an activatory role. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00126 | DAP | Transfer from AT1G06070 | Download |
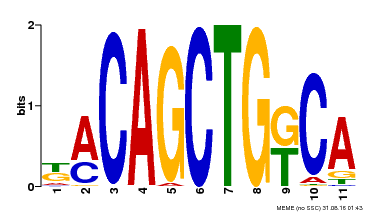
| |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_019168398.1 | 0.0 | PREDICTED: probable transcription factor PosF21 | ||||
| Swissprot | Q04088 | 1e-135 | POF21_ARATH; Probable transcription factor PosF21 | ||||
| TrEMBL | A0A328DWL0 | 0.0 | A0A328DWL0_9ASTE; Uncharacterized protein | ||||
| TrEMBL | A0A484L308 | 0.0 | A0A484L308_9ASTE; Uncharacterized protein | ||||
| STRING | EMJ23579 | 0.0 | (Prunus persica) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Asterids | OGEA3965 | 21 | 37 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G06070.1 | 1e-114 | bZIP family protein | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



