 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Itr_sc001618.1_g00014.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; lamiids; Solanales; Convolvulaceae; Ipomoeeae; Ipomoea
|
||||||||
| Family | G2-like | ||||||||
| Protein Properties | Length: 291aa MW: 31774.6 Da PI: 9.8605 | ||||||||
| Description | G2-like family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | G2-like | 101.6 | 5.2e-32 | 134 | 187 | 2 | 55 |
G2-like 2 prlrWtpeLHerFveaveqLGGsekAtPktilelmkvkgLtlehvkSHLQkYRl 55
pr+rWt++LH++Fv+av+ LGG+e+AtPk++lelm+vk+Ltl+hvkSHLQ+YR+
Itr_sc001618.1_g00014.1 134 PRMRWTSTLHAHFVHAVQLLGGHERATPKSVLELMNVKDLTLAHVKSHLQMYRT 187
9****************************************************7 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF46689 | 1.58E-15 | 131 | 188 | IPR009057 | Homeodomain-like |
| Gene3D | G3DSA:1.10.10.60 | 2.5E-28 | 132 | 188 | IPR009057 | Homeodomain-like |
| TIGRFAMs | TIGR01557 | 6.4E-24 | 134 | 187 | IPR006447 | Myb domain, plants |
| Pfam | PF00249 | 4.5E-7 | 135 | 186 | IPR001005 | SANT/Myb domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0080060 | Biological Process | integument development | ||||
| GO:0005618 | Cellular Component | cell wall | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0044212 | Molecular Function | transcription regulatory region DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 291 aa Download sequence Send to blast |
MSGRGSTLPD LSLQISPPSI SSFDRSCNNI DRSSSMDSGS SGSDLSHEHN GGVSFFPPGY 60 QRSVELSLSR GFELAAAAAG LSGPLSVPVQ RNSRLEVLPQ HRHNFQPQIF GGGGRDFKRS 120 ARMIGGVKRS VRAPRMRWTS TLHAHFVHAV QLLGGHERAT PKSVLELMNV KDLTLAHVKS 180 HLQMYRTVKS TDKAPGARQS CEKAEDMIST PPSSLQTTTP RNAQRASWPP SLPEPNPLYH 240 PHLRQNDAAE MDGRDDGTGR ELCEMKGKFD RLSSSSLSSS SDTFVNLEFT L |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 6j4r_A | 4e-17 | 135 | 189 | 3 | 57 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_B | 4e-17 | 135 | 189 | 3 | 57 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_C | 4e-17 | 135 | 189 | 3 | 57 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_D | 4e-17 | 135 | 189 | 3 | 57 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| Search in ModeBase | ||||||
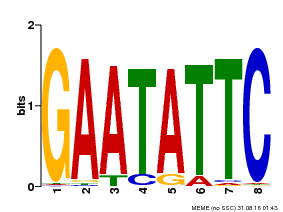
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00107 | PBM | Transfer from AT5G42630 | Download |
| |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_019174419.1 | 1e-160 | PREDICTED: probable transcription factor KAN4 | ||||
| TrEMBL | A0A2G9HEY3 | 3e-71 | A0A2G9HEY3_9LAMI; Uncharacterized protein | ||||
| STRING | XP_006473404.1 | 6e-71 | (Citrus sinensis) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Asterids | OGEA12262 | 18 | 22 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G42630.2 | 3e-50 | G2-like family protein | ||||



