 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Itr_sc003477.1_g00001.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; lamiids; Solanales; Convolvulaceae; Ipomoeeae; Ipomoea
|
||||||||
| Family | bHLH | ||||||||
| Protein Properties | Length: 694aa MW: 74073.2 Da PI: 6.2552 | ||||||||
| Description | bHLH family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | HLH | 13.3 | 0.00015 | 446 | 466 | 35 | 55 |
TS-STCHHHHHHHHHHHHHHH CS
HLH 35 skKlsKaeiLekAveYIksLq 55
s++ +Ka++L +A+eY+k Lq
Itr_sc003477.1_g00001.1 446 SERADKASMLDEAIEYLKTLQ 466
7899****************9 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:4.10.280.10 | 1.3E-4 | 222 | 228 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene3D | G3DSA:4.10.280.10 | 1.3E-4 | 445 | 474 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SuperFamily | SSF47459 | 1.07E-6 | 446 | 494 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| CDD | cd00083 | 8.26E-4 | 446 | 470 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009704 | Biological Process | de-etiolation | ||||
| GO:0009740 | Biological Process | gibberellic acid mediated signaling pathway | ||||
| GO:0010017 | Biological Process | red or far-red light signaling pathway | ||||
| GO:0031539 | Biological Process | positive regulation of anthocyanin metabolic process | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0042802 | Molecular Function | identical protein binding | ||||
| GO:0046983 | Molecular Function | protein dimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 694 aa Download sequence Send to blast |
MPLSEFLKML KNESGERKPT SSSADDLSPL PGSDFSELVW ENGQISMQGQ SSKAKKSDNN 60 LSLQASRMRE KCVGNASTSR IGKLDLAGSV LDEMPPSVPS GEMDLSQDDE IAPWLNYSPT 120 NGQSQEYGSQ LLPEISGMTA NDDGFALIDK GVCCNLMVGA SKSVPNHNVV NSGQRNASKV 180 DCSPTPRFGL LASWSSQQAN PLVSGVSDIG SSNGSINLDS ILKDSVPAQA SAGMKIQNQD 240 TETPRTCPTL LNFSNFSRPV VLARANLQNL TSTLEAKGKR EKEIKENTQN PAKTALIEAC 300 STSRKESELK SQPNLVCTKF EPRPTLAKPH GESSPLEQTD ASFREDTNDA DKAHGKLISP 360 AYVGRVLDGD KTVEPGACSS VCSGSSAERA SNDQSHSLKR RTRENEESGC PSEDAEEESA 420 GAKKAAPARG GTGLKRSRAA EVHNLSERAD KASMLDEAIE YLKTLQLQVQ IMSMGAGLCM 480 PQMMFPAGMH HHMHAQMPHF PQMGLGIGMG MGFGMGLPEL SGRSPGCPIY PMPAMPRPHF 540 PSPSMSGPIN FPGMAAASNI QAFGHPCQGV SMPVPRTPFV PLSQQPSASS APGLNASKMR 600 MNGEVPSKST MLPPEDPEKI KNLQLTQNSD PIPSRLKNQT SQVQTTRDQQ PNASDDPTVN 660 PTNSTNVPGS KAADGLCCAQ RNEIAFIRKG DLAE |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00081 | ChIP-seq | Transfer from AT1G09530 | Download |
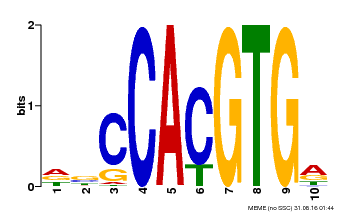
| |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_019170373.1 | 0.0 | PREDICTED: transcription factor PIF3-like isoform X2 | ||||
| TrEMBL | A0A2G3BUP7 | 1e-167 | A0A2G3BUP7_CAPCH; Transcription factor PIF3 (Fragment) | ||||
| STRING | PGSC0003DMT400047079 | 1e-162 | (Solanum tuberosum) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Asterids | OGEA4965 | 24 | 37 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G09530.2 | 3e-31 | phytochrome interacting factor 3 | ||||



