 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Jcr4S00027.250 | ||||||||
| Common Name | JCGZ_23158, LOC105647366 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Malpighiales; Euphorbiaceae; Crotonoideae; Jatropheae; Jatropha
|
||||||||
| Family | MYB | ||||||||
| Protein Properties | Length: 394aa MW: 44298 Da PI: 7.8777 | ||||||||
| Description | MYB family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 58.3 | 1.7e-18 | 14 | 61 | 1 | 48 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHHT CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqkyl 48
+g W++eEde+l+++++++G g+W+++++ g+ R++k+c++rw +yl
Jcr4S00027.250 14 KGLWSPEEDEKLLNYITKHGHGCWSSVPKLAGLQRCGKSCRLRWINYL 61
678*******************************************97 PP
| |||||||
| 2 | Myb_DNA-binding | 45.1 | 2.3e-14 | 67 | 110 | 1 | 46 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHH CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqk 46
rg+++++E+ l++d+++ lG++ W+ Ia+ ++ gRt++++k+ w++
Jcr4S00027.250 67 RGAFSQQEENLIIDLHAVLGNR-WSQIAAQLP-GRTDNEIKNLWNS 110
89********************.*********.*********9986 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:1.10.10.60 | 1.2E-27 | 6 | 64 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS51294 | 25.407 | 9 | 65 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 2.25E-29 | 12 | 108 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 2.2E-13 | 13 | 63 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 2.8E-16 | 14 | 61 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 1.17E-11 | 17 | 61 | No hit | No description |
| Gene3D | G3DSA:1.10.10.60 | 6.4E-25 | 65 | 116 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS51294 | 19.779 | 66 | 116 | IPR017930 | Myb domain |
| SMART | SM00717 | 5.8E-12 | 66 | 114 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 7.0E-13 | 67 | 110 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 1.51E-8 | 69 | 109 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0001944 | Biological Process | vasculature development | ||||
| GO:0009733 | Biological Process | response to auxin | ||||
| GO:0010089 | Biological Process | xylem development | ||||
| GO:0010119 | Biological Process | regulation of stomatal movement | ||||
| GO:0010214 | Biological Process | seed coat development | ||||
| GO:0048364 | Biological Process | root development | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 394 aa Download sequence Send to blast |
MGRHSCCYKQ KLRKGLWSPE EDEKLLNYIT KHGHGCWSSV PKLAGLQRCG KSCRLRWINY 60 LRPDLKRGAF SQQEENLIID LHAVLGNRWS QIAAQLPGRT DNEIKNLWNS CIKKKLRQRG 120 IDPNTHKPLS EVENDHNKEK QTSNIKNNNE LNLIEPAANS KPSSISSSSK ITSNNNNDSS 180 SNLTPPTQEF FIDRYATSSH ETSTNNTTCR PSFVGYFPFQ KLNYRPNNIC FNPNSSSSDM 240 ISDFNSSTII PPSMFQTPIR VKPSVSLPSD NWEASSFSNN GSSCSNGSSS SIELQNNTNF 300 FESNTFSWGL TDCGKSDEED IKWSEYLTST TPFLLGGSVQ NNNQTLYSEV KPETNFITEA 360 SSTTSYHHQQ ASQPCDIYTN KDLQRLAVAF GQTL |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1a5j_A | 6e-28 | 12 | 116 | 5 | 108 | B-MYB |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
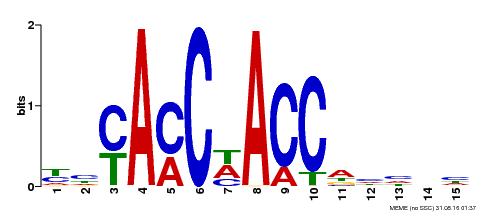
| UniProt | Transcription factor that coordinates a small network of downstream target genes required for several aspects of plant growth and development, such as xylem formation and xylem cell differentiation, and lateral root formation (PubMed:22708996). Regulates a specific set of target genes by binding DNA to the AC cis-element 5'-ACCTAC-3' (PubMed:23741471). Functions as a transcriptional regulator of stomatal closure. Plays a role the regulation of stomatal pore size independently of abscisic acid (ABA) (PubMed:16005292). Required for seed coat mucilage deposition during the development of the seed coat epidermis (PubMed:19401413). Involved in the induction of trichome initiation and branching by positively regulating GL1 and GL2. Required for gibberellin (GA) biosynthesis and degradation by positively affecting the expression of the enzymes that convert GA9 into the bioactive GA4, as well as the enzymes involved in the degradation of GA4 (PubMed:28207974). {ECO:0000269|PubMed:16005292, ECO:0000269|PubMed:19401413, ECO:0000269|PubMed:22708996, ECO:0000269|PubMed:23741471, ECO:0000269|PubMed:28207974}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00134 | DAP | Transfer from AT1G09540 | Download |
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | KM034657 | 0.0 | KM034657.1 Jatropha curcas clone JcMYB037 MYB family protein gene, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_012088808.1 | 0.0 | transcription factor MYB86 | ||||
| Swissprot | Q8VZQ2 | 1e-114 | MYB61_ARATH; Transcription factor MYB61 | ||||
| TrEMBL | A0A067JTK3 | 0.0 | A0A067JTK3_JATCU; MYB family protein | ||||
| STRING | cassava4.1_007846m | 0.0 | (Manihot esculenta) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF1083 | 33 | 103 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G09540.1 | 1e-107 | myb domain protein 61 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Entrez Gene | 105647366 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



