 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Jcr4S00030.150 | ||||||||
| Common Name | JCGZ_04346, LOC105633708 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Malpighiales; Euphorbiaceae; Crotonoideae; Jatropheae; Jatropha
|
||||||||
| Family | G2-like | ||||||||
| Protein Properties | Length: 334aa MW: 37071.5 Da PI: 9.487 | ||||||||
| Description | G2-like family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | G2-like | 101 | 7.5e-32 | 134 | 187 | 2 | 55 |
G2-like 2 prlrWtpeLHerFveaveqLGGsekAtPktilelmkvkgLtlehvkSHLQkYRl 55
pr+rWt++LH++Fv+av+ LGG+e+AtPk++lelm+vk+Ltl+hvkSHLQ+YR+
Jcr4S00030.150 134 PRMRWTTTLHAHFVHAVQLLGGHERATPKSVLELMNVKDLTLAHVKSHLQMYRT 187
9****************************************************7 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF46689 | 1.65E-15 | 131 | 188 | IPR009057 | Homeodomain-like |
| Gene3D | G3DSA:1.10.10.60 | 3.8E-28 | 132 | 188 | IPR009057 | Homeodomain-like |
| TIGRFAMs | TIGR01557 | 2.2E-23 | 134 | 187 | IPR006447 | Myb domain, plants |
| Pfam | PF00249 | 2.1E-7 | 135 | 186 | IPR001005 | SANT/Myb domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0080060 | Biological Process | integument development | ||||
| GO:0005618 | Cellular Component | cell wall | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0044212 | Molecular Function | transcription regulatory region DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 334 aa Download sequence Send to blast |
MRSASTLPDL SLQISPPSAS DCEAKKMAYE GVLSRKALYS DRSSTTDSGS SGSDLSHENG 60 FFSQERSYNL GPSEPTLSLG FEMTDLSHKT LPLPRNLNNH IHNHHHHHQP QIYGREFKRN 120 GRMISGVKRS IRAPRMRWTT TLHAHFVHAV QLLGGHERAT PKSVLELMNV KDLTLAHVKS 180 HLQMYRTVKS TDKGSGQGQT NTGLKQRAGI ADVDALKAEA NPSSLNPPLP PPTSPLATIQ 240 KTQIRGSWSS SMETKDKNRS NTEALAYNSH FNTHETKEDG SMEALQVSDR VKETLERSSL 300 SSSDMLVNLE FTLGRPSWKM DYAESSNELT LLKC |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 6j4k_A | 9e-17 | 135 | 189 | 4 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4k_B | 9e-17 | 135 | 189 | 4 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_A | 9e-17 | 135 | 189 | 4 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_C | 9e-17 | 135 | 189 | 4 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_D | 9e-17 | 135 | 189 | 4 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_F | 9e-17 | 135 | 189 | 4 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_H | 9e-17 | 135 | 189 | 4 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_J | 9e-17 | 135 | 189 | 4 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| Search in ModeBase | ||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00107 | PBM | Transfer from AT5G42630 | Download |
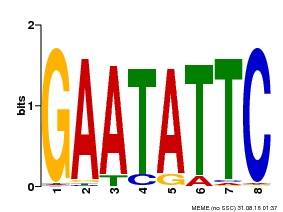
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | - | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_012071737.1 | 0.0 | probable transcription factor KAN4 | ||||
| TrEMBL | A0A067KQD2 | 0.0 | A0A067KQD2_JATCU; Uncharacterized protein | ||||
| STRING | cassava4.1_011255m | 1e-172 | (Manihot esculenta) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF4463 | 33 | 58 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G42630.1 | 2e-74 | G2-like family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Entrez Gene | 105633708 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



