 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Jcr4S00044.40 | ||||||||
| Common Name | JCGZ_11638 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Malpighiales; Euphorbiaceae; Crotonoideae; Jatropheae; Jatropha
|
||||||||
| Family | Dof | ||||||||
| Protein Properties | Length: 471aa MW: 51492 Da PI: 7.047 | ||||||||
| Description | Dof family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | zf-Dof | 127.4 | 4.2e-40 | 120 | 180 | 2 | 62 |
zf-Dof 2 kekalkcprCdstntkfCyynnyslsqPryfCkaCrryWtkGGalrnvPvGggrrknkkss 62
++k+l+cprC+s++tkfCyynny+++qPr+fCkaC+ryWt+GG++rnvPvG+grrknk+s
Jcr4S00044.40 120 PDKILPCPRCNSMDTKFCYYNNYNVNQPRHFCKACQRYWTAGGTMRNVPVGAGRRKNKHSA 180
7899******************************************************986 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| ProDom | PD007478 | 3.0E-30 | 113 | 177 | IPR003851 | Zinc finger, Dof-type |
| Pfam | PF02701 | 1.7E-32 | 122 | 178 | IPR003851 | Zinc finger, Dof-type |
| PROSITE profile | PS50884 | 28.863 | 124 | 178 | IPR003851 | Zinc finger, Dof-type |
| PROSITE pattern | PS01361 | 0 | 126 | 162 | IPR003851 | Zinc finger, Dof-type |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 471 aa Download sequence Send to blast |
MQENNNSNSG SKDSAIKLFG KKIPLKSDSD PPAISCDELP SWERVEEGEA EKTEKDSSAG 60 NVKETSQEDD TEESSDQETN ENPKTPSISE ESAKSKISET EKEQNDPANT TTQEKALKKP 120 DKILPCPRCN SMDTKFCYYN NYNVNQPRHF CKACQRYWTA GGTMRNVPVG AGRRKNKHSA 180 SHYRHITISE ALQAARIEAP NGTHNPALKT NGRVLSFGLD TPICDSMASV LNLADQNVLN 240 GATRNGFHNL EGQKKILGSC KGRENGDDCS SASSVTVSNS VEEGGKICQQ EHIMQNVNGF 300 ASPIQCLPGV PWPYPWNSAL PPPVFCPPGY PMSFYPPFGN YGWNIPLLSP QSSSSSTNQK 360 AQSSPNSPTL GKHSRDGDAL KQDDLEKEES PKRKNGCVLV PKTLRIDDPT EAAKSSIWAT 420 LGIKNETYSS GGLFKSFQSK NNEKNHVNKT SPVLHANPAA LSRSINFHES S |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription factor that binds specifically to a 5'-AA[AG]G-3' consensus core sequence (By similarity). Regulates a photoperiodic flowering response. Transcriptional repressor of 'CONSTANS' expression. {ECO:0000250, ECO:0000269|PubMed:19619493}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00394 | DAP | Transfer from AT3G47500 | Download |
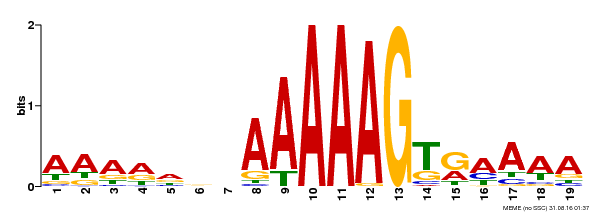
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Circadian-regulation. Highly expressed at the beginning of the light period, then decreases, reaching a minimum between 16 and 29 hours after dawn before rising again at the end of the day. {ECO:0000269|PubMed:19619493}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | GQ256647 | 0.0 | GQ256647.1 Jatropha curcas Dof1 protein mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | NP_001295662.1 | 0.0 | cyclic dof factor 3 | ||||
| Swissprot | Q8LFV3 | 1e-129 | CDF3_ARATH; Cyclic dof factor 3 | ||||
| TrEMBL | A0A067K585 | 0.0 | A0A067K585_JATCU; Uncharacterized protein | ||||
| STRING | cassava4.1_006801m | 0.0 | (Manihot esculenta) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF625 | 34 | 147 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G47500.1 | 1e-113 | cycling DOF factor 3 | ||||



