 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Jcr4S00197.90 | ||||||||
| Common Name | JCGZ_21617, LOC105649716 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Malpighiales; Euphorbiaceae; Crotonoideae; Jatropheae; Jatropha
|
||||||||
| Family | HSF | ||||||||
| Protein Properties | Length: 406aa MW: 46220.2 Da PI: 4.7855 | ||||||||
| Description | HSF family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | HSF_DNA-bind | 110.5 | 1.2e-34 | 14 | 105 | 2 | 102 |
HHHHHHHHHCTGGGTTTSEESSSSSEEEES-HHHHHHHTHHHHSTT--HHHHHHHHHHTTEEE---SSBTTTTXTTSEEEEESXXXXXXXXXXXXX CS
HSF_DNA-bind 2 FlkklyeiledeelkeliswsengnsfvvldeeefakkvLpkyFkhsnfaSFvRQLnmYgFkkvkdeekkskskekiweFkhksFkkgkkelleki 97
Fl+k+ye+++d+++++++sws++++sf+v+ + ef++++Lp++Fkh+nf+SF+RQLn+YgF+k + e+ weF+++ F +gk +l+++i
Jcr4S00197.90 14 FLSKTYEMVDDPSTDSIVSWSQTNKSFIVWSPPEFSRDLLPRFFKHNNFSSFIRQLNTYGFRKADPEQ---------WEFANEDFVRGKPHLMKNI 100
9**************************************************************99988.........******************* PP
XXXXX CS
HSF_DNA-bind 98 krkks 102
+r+k
Jcr4S00197.90 101 HRRKP 105
**985 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:1.10.10.10 | 4.9E-38 | 5 | 98 | IPR011991 | Winged helix-turn-helix DNA-binding domain |
| SuperFamily | SSF46785 | 1.63E-34 | 9 | 103 | IPR011991 | Winged helix-turn-helix DNA-binding domain |
| SMART | SM00415 | 8.5E-62 | 10 | 103 | IPR000232 | Heat shock factor (HSF)-type, DNA-binding |
| Pfam | PF00447 | 1.9E-31 | 14 | 103 | IPR000232 | Heat shock factor (HSF)-type, DNA-binding |
| PRINTS | PR00056 | 1.0E-19 | 14 | 37 | IPR000232 | Heat shock factor (HSF)-type, DNA-binding |
| PRINTS | PR00056 | 1.0E-19 | 52 | 64 | IPR000232 | Heat shock factor (HSF)-type, DNA-binding |
| PRINTS | PR00056 | 1.0E-19 | 65 | 77 | IPR000232 | Heat shock factor (HSF)-type, DNA-binding |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0000302 | Biological Process | response to reactive oxygen species | ||||
| GO:0010200 | Biological Process | response to chitin | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0042803 | Molecular Function | protein homodimerization activity | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 406 aa Download sequence Send to blast |
MDESQGSSNS LPPFLSKTYE MVDDPSTDSI VSWSQTNKSF IVWSPPEFSR DLLPRFFKHN 60 NFSSFIRQLN TYGFRKADPE QWEFANEDFV RGKPHLMKNI HRRKPVHSHS MQNLQGQGSN 120 PLTDSERQAL KDDVDRLKNE KEVLILELQR HDCERQGFEL QMQALKEKLQ QMERRQQTMV 180 SFVSRVLQKP GLALNLMSEL ETGHDRKRRL PRIGYFYEEA SIEDNQMATC QGLANGSADI 240 TSTPLSNTEQ FEQLESSLTF WETTVNDVQT NIQRNSTSEL DESISCGESP AISCVQFNVD 300 VRPKSPAIDM NSEPAVASAP EPDPPKEQVS GTAPTAATGV NDVFWEQFLT ENPGATDAQE 360 VQSERKDSGD RKNEIKPSEQ GKFWWNMRNV NNLAEQMGHL TPAERT |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 5d5u_B | 8e-23 | 4 | 103 | 20 | 129 | Heat shock factor protein 1 |
| 5d5v_B | 8e-23 | 4 | 103 | 20 | 129 | Heat shock factor protein 1 |
| 5d5v_D | 8e-23 | 4 | 103 | 20 | 129 | Heat shock factor protein 1 |
| Search in ModeBase | ||||||
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 204 | 209 | DRKRRL |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcriptional activator that specifically binds DNA sequence 5'-AGAAnnTTCT-3' known as heat shock promoter elements (HSE). | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00443 | DAP | Transfer from AT4G18880 | Download |
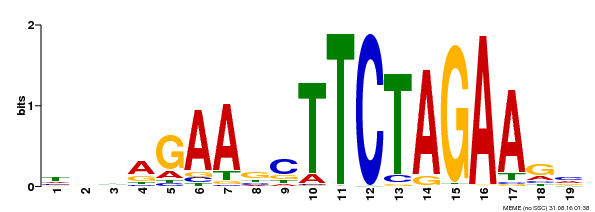
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_012091841.1 | 0.0 | heat stress transcription factor A-4a | ||||
| Swissprot | O49403 | 1e-143 | HFA4A_ARATH; Heat stress transcription factor A-4a | ||||
| TrEMBL | A0A067JBG6 | 0.0 | A0A067JBG6_JATCU; Uncharacterized protein | ||||
| STRING | XP_002522270.1 | 0.0 | (Ricinus communis) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF3568 | 33 | 61 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G18880.1 | 1e-145 | heat shock transcription factor A4A | ||||
| Link Out ? help Back to Top | |
|---|---|
| Entrez Gene | 105649716 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



