 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Jcr4S00433.80 | ||||||||
| Common Name | JCGZ_21170 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Malpighiales; Euphorbiaceae; Crotonoideae; Jatropheae; Jatropha
|
||||||||
| Family | TALE | ||||||||
| Protein Properties | Length: 378aa MW: 42288.7 Da PI: 6.4536 | ||||||||
| Description | TALE family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Homeobox | 29 | 1.9e-09 | 306 | 340 | 21 | 55 |
HSSS--HHHHHHHHHHCTS-HHHHHHHHHHHHHHH CS
Homeobox 21 knrypsaeereeLAkklgLterqVkvWFqNrRake 55
k +yps++++ LA+++gL+++q+ +WF N+R ++
Jcr4S00433.80 306 KWPYPSESQKLALAESTGLDQKQINNWFINQRKRH 340
569*****************************885 PP
| |||||||
| 2 | ELK | 36.7 | 8.9e-13 | 260 | 281 | 1 | 22 |
ELK 1 ELKhqLlrKYsgyLgsLkqEFs 22
ELK qLlr Ysg+Lg+LkqEF+
Jcr4S00433.80 260 ELKGQLLRRYSGCLGNLKQEFM 281
9********************8 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SMART | SM01255 | 7.9E-20 | 108 | 152 | IPR005540 | KNOX1 |
| Pfam | PF03790 | 2.2E-22 | 110 | 148 | IPR005540 | KNOX1 |
| SMART | SM01256 | 9.1E-26 | 157 | 208 | IPR005541 | KNOX2 |
| Pfam | PF03791 | 8.1E-21 | 161 | 207 | IPR005541 | KNOX2 |
| SMART | SM01188 | 1.7E-7 | 260 | 281 | IPR005539 | ELK domain |
| PROSITE profile | PS51213 | 10.319 | 260 | 280 | IPR005539 | ELK domain |
| Pfam | PF03789 | 2.9E-10 | 260 | 281 | IPR005539 | ELK domain |
| PROSITE profile | PS50071 | 12.989 | 280 | 343 | IPR001356 | Homeobox domain |
| SMART | SM00389 | 7.4E-13 | 282 | 347 | IPR001356 | Homeobox domain |
| SuperFamily | SSF46689 | 1.92E-19 | 282 | 356 | IPR009057 | Homeodomain-like |
| Gene3D | G3DSA:1.10.10.60 | 3.9E-28 | 285 | 345 | IPR009057 | Homeodomain-like |
| CDD | cd00086 | 1.86E-12 | 292 | 344 | No hit | No description |
| Pfam | PF05920 | 6.9E-17 | 300 | 339 | IPR008422 | Homeobox KN domain |
| PROSITE pattern | PS00027 | 0 | 318 | 341 | IPR017970 | Homeobox, conserved site |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009934 | Biological Process | regulation of meristem structural organization | ||||
| GO:0048440 | Biological Process | carpel development | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 378 aa Download sequence Send to blast |
MEGGSNSTSC MMAFGDNSNG LCPMMMMPLM TSHPHHHHHH PPPPPPNADS SSNTLFLPLP 60 PPTNHQDQNR NSSSASSMIL DDHNNTTTTT TNTNTSCYFM DSNDGSAASV KAKIMAHPHY 120 HRLLAAYINC QKVGAPPEVV ARLEEACASA AAMGPTGTSC VGEDPALDQF MEAYCEMLTK 180 YEQELSKPFK EAMLFLQRVE CQFKALTVSS PNSGDNNLSL SHCLMRVLVA CGEAGERNGS 240 SEEEVDVNNN FIDPQAEDQE LKGQLLRRYS GCLGNLKQEF MKKRKKGKLP KEARQQLLDW 300 WSRHYKWPYP SESQKLALAE STGLDQKQIN NWFINQRKRH WKPSEDMQFV VMDATHPHYY 360 VDNVLGNPFP MDISPTLL |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 275 | 284 | LKQEFMKKRK |
| 2 | 281 | 285 | KKRKK |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Possible transcription activator involved in early embryonic development. Probably binds to the DNA sequence 5'-TGAC-3'. | |||||
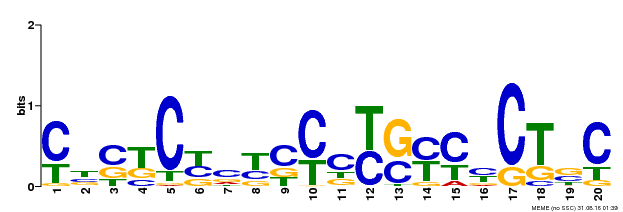
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00675 | ChIP-seq | Transfer from GRMZM2G017087 | Download |
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | EF144909 | 1e-124 | EF144909.1 Populus trichocarpa clone WS01119_M04 unknown mRNA. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_012091289.1 | 0.0 | homeobox protein SBH1 | ||||
| Swissprot | P46608 | 1e-157 | HSBH1_SOYBN; Homeobox protein SBH1 | ||||
| TrEMBL | A0A067JMJ4 | 0.0 | A0A067JMJ4_JATCU; Uncharacterized protein | ||||
| STRING | XP_002518420.1 | 0.0 | (Ricinus communis) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF2039 | 34 | 80 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G62360.1 | 1e-135 | KNOX/ELK homeobox transcription factor | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



