 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Jcr4S01308.20 | ||||||||
| Common Name | JCGZ_09056, LOC105636282 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Malpighiales; Euphorbiaceae; Crotonoideae; Jatropheae; Jatropha
|
||||||||
| Family | Dof | ||||||||
| Protein Properties | Length: 362aa MW: 37605.9 Da PI: 9.0552 | ||||||||
| Description | Dof family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | zf-Dof | 123.5 | 7.2e-39 | 84 | 143 | 3 | 62 |
zf-Dof 3 ekalkcprCdstntkfCyynnyslsqPryfCkaCrryWtkGGalrnvPvGggrrknkkss 62
e+alkcprC+stntkfCy+nnysl+qPr+fCk+CrryWt+GGalrnvPvGgg+r+nk+s+
Jcr4S01308.20 84 ETALKCPRCESTNTKFCYFNNYSLTQPRHFCKTCRRYWTRGGALRNVPVGGGCRRNKRSK 143
7789*****************************************************975 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| ProDom | PD007478 | 5.0E-34 | 67 | 136 | IPR003851 | Zinc finger, Dof-type |
| Pfam | PF02701 | 1.9E-32 | 86 | 141 | IPR003851 | Zinc finger, Dof-type |
| PROSITE profile | PS50884 | 29.544 | 87 | 141 | IPR003851 | Zinc finger, Dof-type |
| PROSITE pattern | PS01361 | 0 | 89 | 125 | IPR003851 | Zinc finger, Dof-type |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0044212 | Molecular Function | transcription regulatory region DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 362 aa Download sequence Send to blast |
MVFSSIPAYL DPANWQQIFV LLSILQQPNN HHQPGGTSGA NTHQLPPPPP PPPLPQPHGS 60 GGAGSIRPGS MADRARLANI PMPETALKCP RCESTNTKFC YFNNYSLTQP RHFCKTCRRY 120 WTRGGALRNV PVGGGCRRNK RSKGSNSKSP VSGDRPTGSG SSSTVSSNSG TSDMLGLVPQ 180 VPPLRFMAPL HHLSEFATGD IGLNYGALSG QVGGPSDLNF QIGSALASVG LGGSGGGCGS 240 GSGSGVGGSL LSVGGLEQWR LHQGPQFPFL GGLDSSSSNS GLYPFESSGG GEPSGYGGGV 300 GQVRPKISSS LTTQFASVKM EDNPELNLSR QLLGMTGNDH YWTGTAWTDL SSFSSSSTSN 360 PL |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00295 | DAP | Transfer from AT2G37590 | Download |
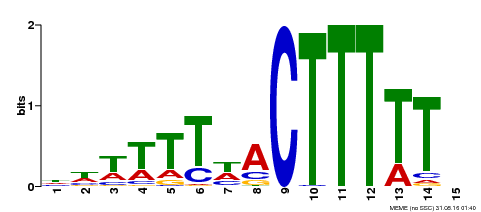
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_012074916.1 | 0.0 | dof zinc finger protein DOF2.4 isoform X1 | ||||
| TrEMBL | A0A067KKT8 | 0.0 | A0A067KKT8_JATCU; Uncharacterized protein | ||||
| STRING | EOY07547 | 1e-150 | (Theobroma cacao) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF656 | 34 | 143 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G37590.1 | 1e-47 | DNA binding with one finger 2.4 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Entrez Gene | 105636282 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



