 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Jcr4S01668.10 | ||||||||
| Common Name | JCGZ_05423 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Malpighiales; Euphorbiaceae; Crotonoideae; Jatropheae; Jatropha
|
||||||||
| Family | C2H2 | ||||||||
| Protein Properties | Length: 371aa MW: 42001.4 Da PI: 8.7903 | ||||||||
| Description | C2H2 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | zf-C2H2 | 22.4 | 3.1e-07 | 197 | 219 | 1 | 23 |
EEETTTTEEESSHHHHHHHHHHT CS
zf-C2H2 1 ykCpdCgksFsrksnLkrHirtH 23
+ C+ Cgk F+r nL+ H+r H
Jcr4S01668.10 197 HYCQVCGKGFKRDANLRMHMRAH 219
68*******************99 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF57667 | 7.4E-6 | 195 | 223 | No hit | No description |
| Gene3D | G3DSA:3.30.160.60 | 1.2E-6 | 197 | 223 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| SMART | SM00355 | 0.0034 | 197 | 219 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE profile | PS50157 | 12.653 | 197 | 224 | IPR007087 | Zinc finger, C2H2 |
| PROSITE pattern | PS00028 | 0 | 199 | 219 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 81 | 258 | 291 | IPR015880 | Zinc finger, C2H2-like |
| SMART | SM00355 | 22 | 296 | 318 | IPR015880 | Zinc finger, C2H2-like |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0010200 | Biological Process | response to chitin | ||||
| GO:0003676 | Molecular Function | nucleic acid binding | ||||
| GO:0046872 | Molecular Function | metal ion binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 371 aa Download sequence Send to blast |
DDTFVSSSIV EASSTMEISS HSKSLLYSLS LLKEKVNQVQ SLVSIIISPD QTAQQPESTS 60 LAITRMSTMI QEIIVTASSM VFTCQQMGTS NPSTNQLHRP AVANANRVQE NELLQSNFGG 120 SDRGKSYFSS ETFDWYGENY NSYNSNRGMV VISNNNNKME NKRENVENNI EEKLSSVMKN 180 YDIIELDAAD LLAKYSHYCQ VCGKGFKRDA NLRMHMRAHG DEYKTHAALS NPMKNSRSEG 240 DDIDNNDKES RLKLPRKYSC PQEGCRWNKK HVKFQPLKSM ICVKNHYKRS HCPKMYVCKR 300 CNRKQFSVLS DLRTHEKHCG DVKWQCSCGT TFSRKDKLMG HVALFIGHSP ATTATSSSTK 360 PATTEMEFKC N |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcription factor. Together with STOP1, plays a critical role in tolerance to major stress factors in acid soils such as proton H(+) and aluminum ion Al(3+). Required for the expression of genes in response to acidic stress (e.g. ALMT1 and MATE), and Al-activated citrate exudation. {ECO:0000269|PubMed:23935008}. | |||||
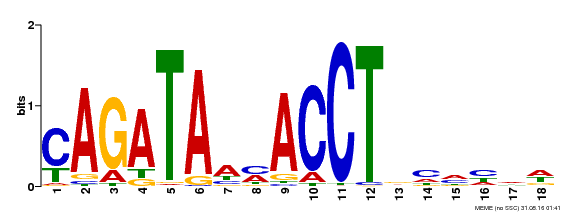
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00522 | DAP | Transfer from AT5G22890 | Download |
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | - | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_020532575.1 | 0.0 | protein SENSITIVE TO PROTON RHIZOTOXICITY 1 | ||||
| Swissprot | Q0WT24 | 1e-114 | STOP2_ARATH; Protein SENSITIVE TO PROTON RHIZOTOXICITY 2 | ||||
| TrEMBL | A0A067L658 | 0.0 | A0A067L658_JATCU; Uncharacterized protein | ||||
| STRING | cassava4.1_031762m | 1e-167 | (Manihot esculenta) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF8099 | 30 | 45 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G22890.1 | 1e-106 | C2H2 family protein | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



