 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Jcr4S02266.20 | ||||||||
| Common Name | JCGZ_06869, LOC105650420 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Malpighiales; Euphorbiaceae; Crotonoideae; Jatropheae; Jatropha
|
||||||||
| Family | NAC | ||||||||
| Protein Properties | Length: 382aa MW: 42787.1 Da PI: 8.4251 | ||||||||
| Description | NAC family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | NAM | 170.3 | 6e-53 | 8 | 143 | 1 | 128 |
NAM 1 lppGfrFhPtdeelvveyLkkkvegkkleleevikevdiykvePwdLpkkvkaeekewyfFskrdkkyatgkrknratksgyWkatgkdkevlsk. 95
lppGfrFhPtdeel+ +yLkkk+++ ++++ ++i++vdiyk++PwdLp k++ +ekewyfFs+rd+ky++g r+nra++sgyWkatg+dk ++++
Jcr4S02266.20 8 LPPGFRFHPTDEELILHYLKKKISSAPFPV-SIIADVDIYKFDPWDLPDKAAFGEKEWYFFSPRDRKYPNGARPNRAAASGYWKATGTDKLIVASs 102
79****************************.89***************888889***************************************999 PP
NAM 96 ........kgelvglkktLvfykgrapkgektdWvmheyrl 128
+++++g+kk Lvfykgr pkg kt+W+mheyrl
Jcr4S02266.20 103 iagssigvQENNIGVKKALVFYKGRPPKGIKTNWIMHEYRL 143
998888888888***************************98 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF101941 | 1.83E-63 | 5 | 179 | IPR003441 | NAC domain |
| PROSITE profile | PS51005 | 60.521 | 8 | 179 | IPR003441 | NAC domain |
| Pfam | PF02365 | 1.2E-27 | 9 | 143 | IPR003441 | NAC domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 382 aa Download sequence Send to blast |
MKNPQSNLPP GFRFHPTDEE LILHYLKKKI SSAPFPVSII ADVDIYKFDP WDLPDKAAFG 60 EKEWYFFSPR DRKYPNGARP NRAAASGYWK ATGTDKLIVA SSIAGSSIGV QENNIGVKKA 120 LVFYKGRPPK GIKTNWIMHE YRLSDTPTHS TIRPINRPKD SSTTTMRLDD WVLCRIYKKT 180 HASPPPAVAA ATTSDQDQED EEEEFVQETL SPSLKNPIIS NKPLMPQKSV SFSNLLDAMD 240 YSMLSSFLSD NQFNPSGYDS TPTLFNNATS SDPFFNNYNP PTANGSGSGG GGGSSSNRFL 300 LQKLPQLNGP LMPNMENKLK RPISTVDQED MHNPSKKIIN SCCFTNSSNQ TDHMPHHHQY 360 NFLNQPFLNQ QLILSPHLQF QG |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 3swm_A | 3e-70 | 5 | 183 | 17 | 172 | NAC domain-containing protein 19 |
| 3swm_B | 3e-70 | 5 | 183 | 17 | 172 | NAC domain-containing protein 19 |
| 3swm_C | 3e-70 | 5 | 183 | 17 | 172 | NAC domain-containing protein 19 |
| 3swm_D | 3e-70 | 5 | 183 | 17 | 172 | NAC domain-containing protein 19 |
| 3swp_A | 3e-70 | 5 | 183 | 17 | 172 | NAC domain-containing protein 19 |
| 3swp_B | 3e-70 | 5 | 183 | 17 | 172 | NAC domain-containing protein 19 |
| 3swp_C | 3e-70 | 5 | 183 | 17 | 172 | NAC domain-containing protein 19 |
| 3swp_D | 3e-70 | 5 | 183 | 17 | 172 | NAC domain-containing protein 19 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription factor that binds to the promoter of ACO5, an ACC oxidase involved in ethylene biosynthesis. Mediates waterlogging-induced hyponastic leaf movement, and cell expansion in abaxial cells of the basal petiole region, by directly regulating the expression of ACO5 (PubMed:24363315). Required for normal seed development and morphology (PubMed:18849494). {ECO:0000269|PubMed:18849494, ECO:0000269|PubMed:24363315}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00333 | DAP | Transfer from AT3G04070 | Download |
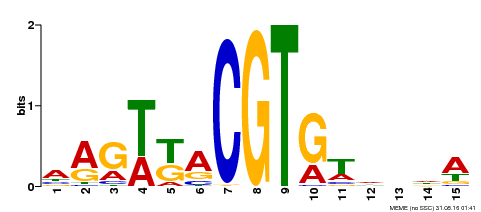
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: By root flooding (PubMed:24363315). Induced by senescence (PubMed:24659488). {ECO:0000269|PubMed:24363315, ECO:0000269|PubMed:24659488}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | KM454464 | 0.0 | KM454464.1 Jatropha curcas NAC047 mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | NP_001306860.1 | 0.0 | NAC transcription factor 29 | ||||
| Swissprot | Q84TD6 | 1e-119 | NAC47_ARATH; NAC transcription factor 47 | ||||
| TrEMBL | R4N5K7 | 0.0 | R4N5K7_JATCU; NAC transcription factor 026 | ||||
| STRING | XP_002525467.1 | 0.0 | (Ricinus communis) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF6621 | 27 | 50 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G04070.1 | 1e-110 | NAC domain containing protein 47 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Entrez Gene | 105650420 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



