 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Jcr4S02405.40 | ||||||||
| Common Name | JCGZ_08042, LOC105635184 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Malpighiales; Euphorbiaceae; Crotonoideae; Jatropheae; Jatropha
|
||||||||
| Family | NAC | ||||||||
| Protein Properties | Length: 368aa MW: 41828.4 Da PI: 5.7925 | ||||||||
| Description | NAC family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | NAM | 171.2 | 3.1e-53 | 21 | 148 | 1 | 128 |
NAM 1 lppGfrFhPtdeelvveyLkkkvegkkleleevikevdiykvePwdLpkkvkaeekewyfFskrdkkyatgkrknratksgyWkatgkdkevlsk. 95
lppGfrFhPtdeel+++yL+ kv + + i+evd++++ePw+Lp +k +e+ewyfFs rd+ky+tg r+nrat +gyWkatgkd+ev+s+
Jcr4S02405.40 21 LPPGFRFHPTDEELITFYLASKVFNGAFCG-VEIAEVDLNRCEPWELPDVAKMGEREWYFFSLRDRKYPTGLRTNRATGAGYWKATGKDREVYSAs 115
79*************************777.45***************8888899**************************************987 PP
NAM 96 kgelvglkktLvfykgrapkgektdWvmheyrl 128
+g+l g+kktLvfykgrap+gekt+Wvmheyrl
Jcr4S02405.40 116 TGALLGMKKTLVFYKGRAPRGEKTKWVMHEYRL 148
8889***************************98 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF101941 | 1.96E-63 | 16 | 171 | IPR003441 | NAC domain |
| PROSITE profile | PS51005 | 59.837 | 21 | 170 | IPR003441 | NAC domain |
| Pfam | PF02365 | 8.8E-28 | 22 | 148 | IPR003441 | NAC domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0010014 | Biological Process | meristem initiation | ||||
| GO:0010199 | Biological Process | organ boundary specification between lateral organs and the meristem | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 368 aa Download sequence Send to blast |
MLSMEELLCE LIGEDKNEQG LPPGFRFHPT DEELITFYLA SKVFNGAFCG VEIAEVDLNR 60 CEPWELPDVA KMGEREWYFF SLRDRKYPTG LRTNRATGAG YWKATGKDRE VYSASTGALL 120 GMKKTLVFYK GRAPRGEKTK WVMHEYRLDG DFSYRHTCKE EWVICRIFHK NGEKKNGILQ 180 AQGFLLEANS PPTFSSLPSL LEAPLLLECE TQTPFLIKQD SQSNKHTVSF GTSSPITIST 240 DAETNKNIQD NPSAPMLFKS LLSHPLKEQF TIPKQCKTEP NFSHFQLPAG ADHANLFNYW 300 VEKIHHPNTY HCPLSFEMDC NVLGISTEFD SDITANDLST SIAFNRADFQ MMLDTPIRLP 360 AQSWPLDS |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1ut4_A | 1e-50 | 21 | 171 | 17 | 166 | NO APICAL MERISTEM PROTEIN |
| 1ut4_B | 1e-50 | 21 | 171 | 17 | 166 | NO APICAL MERISTEM PROTEIN |
| 1ut7_A | 1e-50 | 21 | 171 | 17 | 166 | NO APICAL MERISTEM PROTEIN |
| 1ut7_B | 1e-50 | 21 | 171 | 17 | 166 | NO APICAL MERISTEM PROTEIN |
| 3swm_A | 1e-50 | 21 | 171 | 20 | 169 | NAC domain-containing protein 19 |
| 3swm_B | 1e-50 | 21 | 171 | 20 | 169 | NAC domain-containing protein 19 |
| 3swm_C | 1e-50 | 21 | 171 | 20 | 169 | NAC domain-containing protein 19 |
| 3swm_D | 1e-50 | 21 | 171 | 20 | 169 | NAC domain-containing protein 19 |
| 3swp_A | 1e-50 | 21 | 171 | 20 | 169 | NAC domain-containing protein 19 |
| 3swp_B | 1e-50 | 21 | 171 | 20 | 169 | NAC domain-containing protein 19 |
| 3swp_C | 1e-50 | 21 | 171 | 20 | 169 | NAC domain-containing protein 19 |
| 3swp_D | 1e-50 | 21 | 171 | 20 | 169 | NAC domain-containing protein 19 |
| 4dul_A | 1e-50 | 21 | 171 | 17 | 166 | NAC domain-containing protein 19 |
| 4dul_B | 1e-50 | 21 | 171 | 17 | 166 | NAC domain-containing protein 19 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription activator. Involved in molecular mechanisms regulating shoot apical meristem (SAM) formation during embryogenesis and organ separation. Required for axillary meristem initiation and separation of the meristem from the main stem. May act as an inhibitor of cell division. {ECO:0000269|PubMed:12837947, ECO:0000269|PubMed:17122068}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00241 | DAP | Transfer from AT1G76420 | Download |
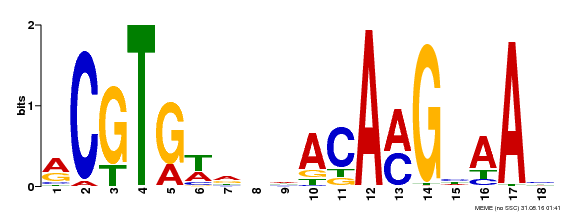
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: By BRM, at the chromatin level, and conferring a very specific spatial expression pattern. {ECO:0000269|PubMed:16854978}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | KC775285 | 0.0 | KC775285.1 Jatropha curcas NAC transcription factor 007 gene, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_020535452.1 | 0.0 | protein CUP-SHAPED COTYLEDON 3 isoform X2 | ||||
| Swissprot | Q9S851 | 1e-105 | NAC31_ARATH; Protein CUP-SHAPED COTYLEDON 3 | ||||
| TrEMBL | R4N7J5 | 0.0 | R4N7J5_JATCU; NAC transcription factor 007 | ||||
| STRING | XP_002510719.1 | 0.0 | (Ricinus communis) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF7888 | 33 | 47 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G76420.1 | 1e-107 | NAC family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Entrez Gene | 105635184 |



