 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Jcr4S04680.20 | ||||||||
| Common Name | GRAS36, JCGZ_04722, LOC105634083 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Malpighiales; Euphorbiaceae; Crotonoideae; Jatropheae; Jatropha
|
||||||||
| Family | GRAS | ||||||||
| Protein Properties | Length: 623aa MW: 67941.4 Da PI: 5.1847 | ||||||||
| Description | GRAS family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | GRAS | 459.7 | 1.8e-140 | 260 | 619 | 1 | 374 |
GRAS 1 lvelLlecAeavssgdlelaqalLarlselaspdgdpmqRlaayfteALaarlarsvselykalppsetseknsseelaalklfsevsPilkfshl 96
lv+lL++cAeav++++l+la+al++++ la++++ +m+++a+yf+eALa+r++r ly pps+ + ++ s+ l++ f+e++P+lkf+h+
Jcr4S04680.20 260 LVHLLMACAEAVQDNNLTLAEALVKQIGFLAASQAGAMRKVATYFAEALARRIYR----LY---PPSPVD-HSLSDILQM--HFYETCPYLKFAHF 345
68*****************************************************....44...555554.345555444..5************* PP
GRAS 97 taNqaIleavegeervHiiDfdisqGlQWpaLlqaLasRpegppslRiTgvgspesgskeeleetgerLakfAeelgvpfefnvlvakrledlele 192
taNqaIlea+eg++rvH+iDf++++G+QWpaLlqaLa Rp+gpp +R+Tg+g+p++++++ l+e+g++La++Ae+++v+fe++ +va++l+dl+ +
Jcr4S04680.20 346 TANQAILEAFEGKKRVHVIDFSMNEGMQWPALLQALALRPGGPPAFRLTGIGPPSHDDSDHLQEVGWKLAQLAETIHVEFEYRGFVANSLADLDAS 441
************************************************************************************************ PP
GRAS 193 eLrvkp..gEalaVnlvlqlhrlldesvsleserdevLklvkslsPkvvvvveqeadhnsesFlerflealeyysalfdsleaklpreseerikvE 286
+L+++p E+laVn+v++lh+ll++++++e+ vL++vk+++P++v++veqea+hn++ Fl+rf+e+l+yys+lfdsle + +s ++++++
Jcr4S04680.20 442 MLELRPneFESLAVNSVFELHKLLARPGAIEK----VLSVVKQMKPEIVTIVEQEANHNGPVFLDRFTESLHYYSTLFDSLEGS---ASTQDKVMS 530
******77789*********************....***********************************************9...6999***** PP
GRAS 287 rellgreivnvvacegaerrerhetlekWrerleeaGFkpvplsekaakqaklllrkvk.sdgyrveeesgslvlgWkdrpLvsvSaWr 374
+++lg++i+nvvacega+r+erhetl +Wr rl+++GF pv+l+++a kqa++ll+ ++ +dgyrvee++g+l+lgW++rpL+++SaWr
Jcr4S04680.20 531 EVYLGKQICNVVACEGADRVERHETLTQWRTRLGSSGFVPVHLGSNAFKQASMLLALFAgGDGYRVEENNGCLTLGWHTRPLIATSAWR 619
****************************************************************************************8 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SMART | SM01129 | 8.3E-40 | 54 | 127 | No hit | No description |
| Pfam | PF12041 | 2.6E-37 | 54 | 121 | IPR021914 | Transcriptional factor DELLA, N-terminal |
| PROSITE profile | PS50985 | 68.192 | 234 | 598 | IPR005202 | Transcription factor GRAS |
| Pfam | PF03514 | 6.2E-138 | 260 | 619 | IPR005202 | Transcription factor GRAS |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009723 | Biological Process | response to ethylene | ||||
| GO:0009737 | Biological Process | response to abscisic acid | ||||
| GO:0009867 | Biological Process | jasmonic acid mediated signaling pathway | ||||
| GO:0009938 | Biological Process | negative regulation of gibberellic acid mediated signaling pathway | ||||
| GO:0010187 | Biological Process | negative regulation of seed germination | ||||
| GO:0010218 | Biological Process | response to far red light | ||||
| GO:0042176 | Biological Process | regulation of protein catabolic process | ||||
| GO:0042538 | Biological Process | hyperosmotic salinity response | ||||
| GO:2000033 | Biological Process | regulation of seed dormancy process | ||||
| GO:2000377 | Biological Process | regulation of reactive oxygen species metabolic process | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0044212 | Molecular Function | transcription regulatory region DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 623 aa Download sequence Send to blast |
MKREHSNQHL RLDPTSAGSS STAYSSMAAT SAAPDTSCKS KIWEDEAQGD GGMDELLAVL 60 GYKVRSSDMA EVAQKLEQLE EVMCNVQEDG LSHLSSETVH YNPSDISTWL ESMLSELNPN 120 PNFDPQPSLN DSFFAAPESS TVTSIDFAEQ QKSNKHASGV MAFEESSSDY DLKAIPGKAV 180 FARSHQIDSS SSSPRDPKRL KPTYPPLPPP PPAAAAATSS SSSTISGVSV SGSGSVSFGV 240 STESTRPVVL VDSQENGIRL VHLLMACAEA VQDNNLTLAE ALVKQIGFLA ASQAGAMRKV 300 ATYFAEALAR RIYRLYPPSP VDHSLSDILQ MHFYETCPYL KFAHFTANQA ILEAFEGKKR 360 VHVIDFSMNE GMQWPALLQA LALRPGGPPA FRLTGIGPPS HDDSDHLQEV GWKLAQLAET 420 IHVEFEYRGF VANSLADLDA SMLELRPNEF ESLAVNSVFE LHKLLARPGA IEKVLSVVKQ 480 MKPEIVTIVE QEANHNGPVF LDRFTESLHY YSTLFDSLEG SASTQDKVMS EVYLGKQICN 540 VVACEGADRV ERHETLTQWR TRLGSSGFVP VHLGSNAFKQ ASMLLALFAG GDGYRVEENN 600 GCLTLGWHTR PLIATSAWRV VNK |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 5b3g_A | 2e-66 | 255 | 618 | 14 | 378 | Protein SCARECROW |
| 5b3h_A | 2e-66 | 255 | 618 | 13 | 377 | Protein SCARECROW |
| 5b3h_D | 2e-66 | 255 | 618 | 13 | 377 | Protein SCARECROW |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcriptional regulator that acts as a repressor of the gibberellin (GA) signaling pathway. Probably acts by participating in large multiprotein complexes that repress transcription of GA-inducible genes. Upon GA application, it is degraded by the proteasome, allowing the GA signaling pathway. | |||||
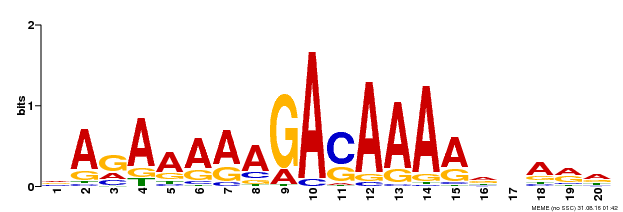
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00611 | ChIP-seq | Transfer from AT2G01570 | Download |
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | KM405268 | 0.0 | KM405268.1 Jatropha curcas GRAS36 protein (GRAS36) mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_012072251.1 | 0.0 | DELLA protein GAIP-B | ||||
| Swissprot | Q8S4W7 | 0.0 | GAI1_VITVI; DELLA protein GAI1 | ||||
| TrEMBL | A0A067L1V6 | 0.0 | A0A067L1V6_JATCU; GRAS36 protein | ||||
| STRING | cassava4.1_033968m | 0.0 | (Manihot esculenta) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF803 | 34 | 121 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G01570.1 | 0.0 | GRAS family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Entrez Gene | 105634083 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



