 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Jcr4S04809.10 | ||||||||
| Common Name | JCGZ_01467, LOC105647749 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Malpighiales; Euphorbiaceae; Crotonoideae; Jatropheae; Jatropha
|
||||||||
| Family | Dof | ||||||||
| Protein Properties | Length: 338aa MW: 35691.7 Da PI: 9.1212 | ||||||||
| Description | Dof family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | zf-Dof | 124.2 | 4.4e-39 | 69 | 129 | 2 | 62 |
zf-Dof 2 kekalkcprCdstntkfCyynnyslsqPryfCkaCrryWtkGGalrnvPvGggrrknkkss 62
+e+alkcprC+stntkfCy+nnyslsqPr+fCk+CrryWt+GGalrnvPvGgg+r+nk+s+
Jcr4S04809.10 69 PEAALKCPRCESTNTKFCYFNNYSLSQPRHFCKTCRRYWTRGGALRNVPVGGGCRRNKRSK 129
67899*****************************************************975 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| ProDom | PD007478 | 2.0E-32 | 53 | 118 | IPR003851 | Zinc finger, Dof-type |
| Pfam | PF02701 | 7.4E-33 | 72 | 127 | IPR003851 | Zinc finger, Dof-type |
| PROSITE profile | PS50884 | 29.609 | 73 | 127 | IPR003851 | Zinc finger, Dof-type |
| PROSITE pattern | PS01361 | 0 | 75 | 111 | IPR003851 | Zinc finger, Dof-type |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009640 | Biological Process | photomorphogenesis | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0044212 | Molecular Function | transcription regulatory region DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 338 aa Download sequence Send to blast |
MVFSSIPVYL DPPNWQQHNQ QSAVSSESHQ LPLPAPPPGC GGGGVSGSAA SIRPGSMSER 60 ARLAKIPQPE AALKCPRCES TNTKFCYFNN YSLSQPRHFC KTCRRYWTRG GALRNVPVGG 120 GCRRNKRSKG NNNNNRSKSP TKAGSASSNG LITNTCTTDN ITGHIIAPPP HLPILPPLHH 180 LNNYNSTDIG LNFGGIQPPV AATAGGGMEF HQLGSTSSAS GAGGGSGGGV GGGCGSFLSN 240 GIVDQWRLQQ VQQFPFMANL GASNGLYQFE NEGINYAGQG RSKPLESGVN DLAAVKMEEM 300 NLSKNFLGIS GNDHQYWSGN NAWTDLSGFT SSSTSHLL |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00407 | DAP | Transfer from AT3G55370 | Download |
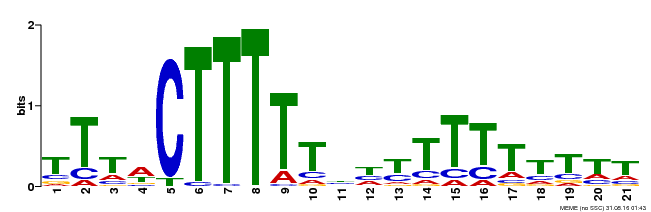
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_012089351.1 | 0.0 | dof zinc finger protein DOF3.6 | ||||
| TrEMBL | A0A067L975 | 0.0 | A0A067L975_JATCU; Uncharacterized protein | ||||
| STRING | cassava4.1_011714m | 1e-130 | (Manihot esculenta) | ||||
| STRING | cassava4.1_011891m | 1e-130 | (Manihot esculenta) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF656 | 34 | 143 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G37590.1 | 8e-42 | DNA binding with one finger 2.4 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Entrez Gene | 105647749 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



