 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Jcr4S04869.20 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Malpighiales; Euphorbiaceae; Crotonoideae; Jatropheae; Jatropha
|
||||||||
| Family | NAC | ||||||||
| Protein Properties | Length: 492aa MW: 54275.6 Da PI: 4.457 | ||||||||
| Description | NAC family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | NAM | 141.6 | 4.6e-44 | 10 | 128 | 1 | 128 |
NAM 1 lppGfrFhPtdeelvveyLkkkvegkkleleevikevdiykvePwdLpkkvkaeekewyfFskrdkkyatgkrknratksgyWkatgkdkevlskk 96
l+pGfrFhPtdeelv +yLk+kv++k++++ + i+ +diyk+ePwdLp k +++ w + +ky +g+++nrat++gyWk+tgkd+++ +
Jcr4S04869.20 10 LAPGFRFHPTDEELVRYYLKRKVTNKPFRF-DPIAVIDIYKSEPWDLPGK---NKRIW----GNFRKYGNGSKTNRATEKGYWKTTGKDRPIQW-N 96
579**************************9.99***************33...34555....445789*************************9.9 PP
NAM 97 gelvglkktLvfykgrapkgektdWvmheyrl 128
+++vg+kktLv++ grap+ge+t+Wvmheyrl
Jcr4S04869.20 97 SRTVGMKKTLVYHLGRAPRGERTNWVMHEYRL 128
99****************************98 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF101941 | 1.01E-53 | 8 | 151 | IPR003441 | NAC domain |
| PROSITE profile | PS51005 | 50.276 | 10 | 151 | IPR003441 | NAC domain |
| Pfam | PF02365 | 1.4E-24 | 12 | 128 | IPR003441 | NAC domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009819 | Biological Process | drought recovery | ||||
| GO:0010150 | Biological Process | leaf senescence | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:2000377 | Biological Process | regulation of reactive oxygen species metabolic process | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0005737 | Cellular Component | cytoplasm | ||||
| GO:0005886 | Cellular Component | plasma membrane | ||||
| GO:0016021 | Cellular Component | integral component of membrane | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 492 aa Download sequence Send to blast |
MVRGDSSTSL APGFRFHPTD EELVRYYLKR KVTNKPFRFD PIAVIDIYKS EPWDLPGKNK 60 RIWGNFRKYG NGSKTNRATE KGYWKTTGKD RPIQWNSRTV GMKKTLVYHL GRAPRGERTN 120 WVMHEYRLAD EDLQKAGIVQ DSFVLCRIFQ KSGTGPKNGE QYGAPFVEEE WDDDEVVALL 180 PGEEMVVTDE VAVGDDACVE ANDLDQNFDG IASGNASVPV NYYHGEASNY VEQSSGNCSG 240 DDQKPIIIGK EETQYGSNLP DDQNLFNLAG QYEVDAKAST GDSTPFDMVD EYLSFFDAND 300 DNLTFDPSEL MGTEYAIPDQ DHLFEKDVNG GAKAVPVVTS ELSEAHGNND ASSSKQLKPE 360 ATMFESDMKY PXXXMLGSIP APPAFASEFP VKDRALQLNA AQSSSSIRVT AGVIRIENIT 420 FGSSGMDWSS NKNQNVNIIL SFDMAQANVG PASLVPMGSF FSGKAMSTVS RGCQGSVIFG 480 VFMVWVGLVD WV |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1ut4_A | 1e-40 | 6 | 157 | 13 | 171 | NO APICAL MERISTEM PROTEIN |
| 1ut4_B | 1e-40 | 6 | 157 | 13 | 171 | NO APICAL MERISTEM PROTEIN |
| 1ut7_A | 1e-40 | 6 | 157 | 13 | 171 | NO APICAL MERISTEM PROTEIN |
| 1ut7_B | 1e-40 | 6 | 157 | 13 | 171 | NO APICAL MERISTEM PROTEIN |
| 3swm_A | 1e-40 | 6 | 157 | 16 | 174 | NAC domain-containing protein 19 |
| 3swm_B | 1e-40 | 6 | 157 | 16 | 174 | NAC domain-containing protein 19 |
| 3swm_C | 1e-40 | 6 | 157 | 16 | 174 | NAC domain-containing protein 19 |
| 3swm_D | 1e-40 | 6 | 157 | 16 | 174 | NAC domain-containing protein 19 |
| 3swp_A | 1e-40 | 6 | 157 | 16 | 174 | NAC domain-containing protein 19 |
| 3swp_B | 1e-40 | 6 | 157 | 16 | 174 | NAC domain-containing protein 19 |
| 3swp_C | 1e-40 | 6 | 157 | 16 | 174 | NAC domain-containing protein 19 |
| 3swp_D | 1e-40 | 6 | 157 | 16 | 174 | NAC domain-containing protein 19 |
| 4dul_A | 1e-40 | 6 | 157 | 13 | 171 | NAC domain-containing protein 19 |
| 4dul_B | 1e-40 | 6 | 157 | 13 | 171 | NAC domain-containing protein 19 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcriptional activator activated by proteolytic cleavage through regulated intramembrane proteolysis (RIP) (Ref.6, PubMed:24323506, PubMed:25219309). Promotes reactive oxygen species (ROS) production during drought-induced leaf senescence. In response to abscisic acid (ABA)-mediated drought stress signals, binds directly to the promoters of RBOHC and RBOHE genes, encoding ROS biosynthetic enzymes, resulting in ROS accumulation and triggering leaf senescence via programmed cell death (PCD). ROS-induced leaf senescence sustains plant survival under drought conditions (PubMed:22313226). Involved in heat stress response. Modulates PCD through a ROS-mediated positive feedback control under heat stress conditions. This may provide an adaptation strategy for plant survival under extreme heat stress conditions (PubMed:25219309). Acts as repressor in preventing anther dehiscence during stamen development by suppressing genes that participate in jasmonic acid (JA) biosynthesis, such as DAD1, AOS, AOC3, OPR3 and 4CLL5/OPCL1 (PubMed:24323506). {ECO:0000269|PubMed:22313226, ECO:0000269|PubMed:24323506, ECO:0000269|PubMed:25219309, ECO:0000269|Ref.6}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00343 | DAP | Transfer from AT3G10500 | Download |
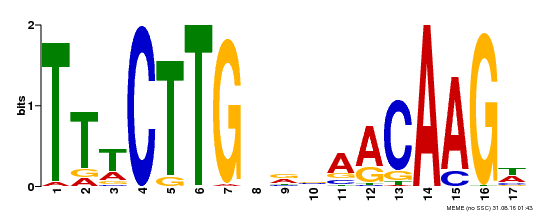
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: By abscisic acid (ABA) (PubMed:22313226, PubMed:25219309). Induced by heat shock (PubMed:25219309). Induced by cold, drought stress and methyl methanesulfonate (MMS) treatment (PubMed:17158162). {ECO:0000269|PubMed:17158162, ECO:0000269|PubMed:22313226, ECO:0000269|PubMed:25219309}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | KC775322 | 1e-165 | KC775322.1 Jatropha curcas NAC transcription factor 044 gene, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_012090539.2 | 0.0 | NAC domain-containing protein 78 | ||||
| Swissprot | Q949N0 | 1e-132 | NAC53_ARATH; NAC domain-containing protein 53 | ||||
| TrEMBL | R4NHQ2 | 0.0 | R4NHQ2_JATCU; NAC transcription factor 044 | ||||
| STRING | cassava4.1_004618m | 0.0 | (Manihot esculenta) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF5347 | 32 | 47 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G10500.1 | 1e-132 | NAC domain containing protein 53 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



