 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Jcr4S05581.10 | ||||||||
| Common Name | JCGZ_02456, LOC105629779 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Malpighiales; Euphorbiaceae; Crotonoideae; Jatropheae; Jatropha
|
||||||||
| Family | C2H2 | ||||||||
| Protein Properties | Length: 287aa MW: 33373 Da PI: 8.6108 | ||||||||
| Description | C2H2 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | zf-C2H2 | 18.5 | 5.4e-06 | 12 | 36 | 1 | 23 |
EEET..TTTEEESSHHHHHHHHHHT CS
zf-C2H2 1 ykCp..dCgksFsrksnLkrHirtH 23
y+C dC+ s++rk++L+rH+ +H
Jcr4S05581.10 12 YVCLvdDCHASYRRKDHLTRHLLKH 36
899999*****************99 PP
| |||||||
| 2 | zf-C2H2 | 15.5 | 5.1e-05 | 41 | 66 | 1 | 23 |
EEET..TTTEEESSHHHHHHHHHH.T CS
zf-C2H2 1 ykCp..dCgksFsrksnLkrHirt.H 23
+kCp +C++ F + n krH + H
Jcr4S05581.10 41 FKCPieNCDRQFVFQGNVKRHVNEfH 66
89*******************99888 PP
| |||||||
| 3 | zf-C2H2 | 19.6 | 2.5e-06 | 80 | 104 | 1 | 23 |
EEET..TTTEEESSHHHHHHHHHHT CS
zf-C2H2 1 ykCp..dCgksFsrksnLkrHirtH 23
y+C+ Cgk+F+ s L++H +H
Jcr4S05581.10 80 YVCQesGCGKVFRYLSKLQKHEDSH 104
99*******************9888 PP
| |||||||
| 4 | zf-C2H2 | 14.4 | 0.00011 | 172 | 196 | 2 | 23 |
EET..TTTEEESSHHHHHHHHHH.T CS
zf-C2H2 2 kCp..dCgksFsrksnLkrHirt.H 23
kC+ C Fs+k nL++H + H
Jcr4S05581.10 172 KCHfeGCRNAFSNKTNLNQHVKAaH 196
7*******************99877 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF57667 | 4.17E-6 | 9 | 38 | No hit | No description |
| Gene3D | G3DSA:3.30.160.60 | 5.0E-9 | 9 | 38 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| SMART | SM00355 | 0.019 | 12 | 36 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE profile | PS50157 | 9.078 | 12 | 41 | IPR007087 | Zinc finger, C2H2 |
| PROSITE pattern | PS00028 | 0 | 14 | 36 | IPR007087 | Zinc finger, C2H2 |
| SuperFamily | SSF57667 | 2.55E-6 | 40 | 67 | No hit | No description |
| SMART | SM00355 | 0.14 | 41 | 66 | IPR015880 | Zinc finger, C2H2-like |
| Gene3D | G3DSA:3.30.160.60 | 5.2E-5 | 41 | 69 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| PROSITE profile | PS50157 | 11.177 | 41 | 71 | IPR007087 | Zinc finger, C2H2 |
| PROSITE pattern | PS00028 | 0 | 43 | 66 | IPR007087 | Zinc finger, C2H2 |
| SuperFamily | SSF57667 | 1.54E-6 | 75 | 106 | No hit | No description |
| Gene3D | G3DSA:3.30.160.60 | 5.2E-8 | 76 | 104 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| SMART | SM00355 | 0.0017 | 80 | 104 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE profile | PS50157 | 10.887 | 80 | 109 | IPR007087 | Zinc finger, C2H2 |
| PROSITE pattern | PS00028 | 0 | 82 | 104 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 5.5 | 112 | 137 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE pattern | PS00028 | 0 | 114 | 137 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 14 | 140 | 161 | IPR015880 | Zinc finger, C2H2-like |
| SuperFamily | SSF57667 | 4.72E-7 | 151 | 208 | No hit | No description |
| Gene3D | G3DSA:3.30.160.60 | 3.0E-6 | 151 | 193 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| SMART | SM00355 | 0.015 | 171 | 196 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE profile | PS50157 | 9.743 | 171 | 201 | IPR007087 | Zinc finger, C2H2 |
| PROSITE pattern | PS00028 | 0 | 173 | 196 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 6.4 | 202 | 228 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE pattern | PS00028 | 0 | 204 | 228 | IPR007087 | Zinc finger, C2H2 |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0005730 | Cellular Component | nucleolus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0008097 | Molecular Function | 5S rRNA binding | ||||
| GO:0046872 | Molecular Function | metal ion binding | ||||
| GO:0080084 | Molecular Function | 5S rDNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 287 aa Download sequence Send to blast |
MRFEGFFIQR PYVCLVDDCH ASYRRKDHLT RHLLKHEGRL FKCPIENCDR QFVFQGNVKR 60 HVNEFHSQKS PSTDVGEKKY VCQESGCGKV FRYLSKLQKH EDSHVKLDSV EAICAEPGCM 120 KHFSDVQCLQ AHVQSCHTYM TCEICGTKQL KKNLKRHLRT HESGGGSMER IKCHFEGCRN 180 AFSNKTNLNQ HVKAAHLEVR RFTCGVPGCD MRFAYKHVRD RHEKSACHVY TAGDFEDFDD 240 QFRSRPRGGR KRECPTVDIL LRKRVLPPSD LDECCSWFQS IGSQEQL |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 2i13_A | 2e-11 | 9 | 194 | 19 | 183 | Aart |
| 2i13_B | 2e-11 | 9 | 194 | 19 | 183 | Aart |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Essential protein (PubMed:22353599). Isoform 1 is a transcription activator the binds both 5S rDNA and 5S rRNA and stimulates the transcription of 5S rRNA gene (PubMed:12711688, PubMed:22353599). Isoform 1 regulates 5S rRNA levels during development (PubMed:22353599). {ECO:0000269|PubMed:12711688, ECO:0000269|PubMed:22353599}. | |||||
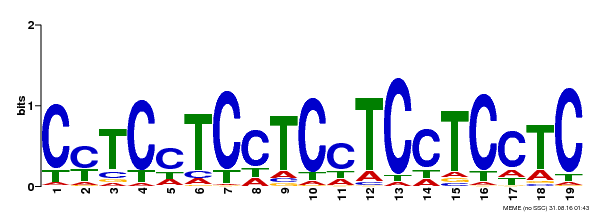
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00229 | DAP | Transfer from AT1G72050 | Download |
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_012066810.1 | 0.0 | transcription factor IIIA | ||||
| Swissprot | Q84MZ4 | 1e-106 | TF3A_ARATH; Transcription factor IIIA | ||||
| TrEMBL | A0A067LE81 | 0.0 | A0A067LE81_JATCU; Uncharacterized protein | ||||
| STRING | cassava4.1_010279m | 1e-160 | (Manihot esculenta) | ||||
| STRING | XP_002533583.1 | 1e-160 | (Ricinus communis) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF3069 | 33 | 70 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G72050.1 | 1e-109 | transcription factor IIIA | ||||
| Link Out ? help Back to Top | |
|---|---|
| Entrez Gene | 105629779 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



