 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Jcr4S06887.40 | ||||||||
| Common Name | JCGZ_12960 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Malpighiales; Euphorbiaceae; Crotonoideae; Jatropheae; Jatropha
|
||||||||
| Family | TCP | ||||||||
| Protein Properties | Length: 354aa MW: 39920.2 Da PI: 9.6907 | ||||||||
| Description | TCP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | TCP | 142.6 | 3.1e-44 | 109 | 246 | 2 | 133 |
TCP 2 agkkdrhskihTkvggRdRRvRlsaecaarfFdLqdeLGfdkdsktieWLlqqakpaikeltgtssssasec..eaesssssasnsssg....kaa 91
+ kkdrhskihT++g+RdRRvRls+++a++fFdLqd+LGfdk+skt+eWLl ++k+aik + ss++s++ ++ + s +n + +
Jcr4S06887.40 109 PLKKDRHSKIHTSQGLRDRRVRLSIQIARKFFDLQDMLGFDKASKTLEWLLSKSKRAIKRVAHKRSSPSSSAssSSTCEALSPENG--EiameEGI 202
679*************************************************************7666644422112222222222..24466333 PP
TCP 92 ksaa.kskksqksaasalnlak.esrakarararertrekmrik 133
s+ ++kk++k ++ + nla+ e r karararertr km ++
Jcr4S06887.40 203 VSTGkNEKKAKKLEKYTTNLAAkELRVKARARARERTRIKMLNQ 246
33333566666666666676666******************887 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51369 | 34.852 | 111 | 169 | IPR017887 | Transcription factor TCP subgroup |
| Pfam | PF03634 | 6.2E-41 | 111 | 243 | IPR005333 | Transcription factor, TCP |
| PROSITE profile | PS51370 | 10.643 | 225 | 242 | IPR017888 | CYC/TB1, R domain |
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 354 aa Download sequence Send to blast |
MFPSTINATI NPFPYLSSFS SYNPSPFFLN NHEATTSTDV FFQPTFSLLP NNPLTSSLVS 60 TETLSSINAK QDINCYNIDQ IDGESCYKQS QQLQQQQPSS CVFVSAKKPL KKDRHSKIHT 120 SQGLRDRRVR LSIQIARKFF DLQDMLGFDK ASKTLEWLLS KSKRAIKRVA HKRSSPSSSA 180 SSSSTCEALS PENGEIAMEE GIVSTGKNEK KAKKLEKYTT NLAAKELRVK ARARARERTR 240 IKMLNQYLLS NDELVDELPD AKENNIIEES IAIERKLKPS SVMGYQQNLL MLRDLTCNNS 300 NYNNLNLPNF TSNWEINNAI ARSSFCAITN MNRSAGLHLY GKLREAENGS QCLH |
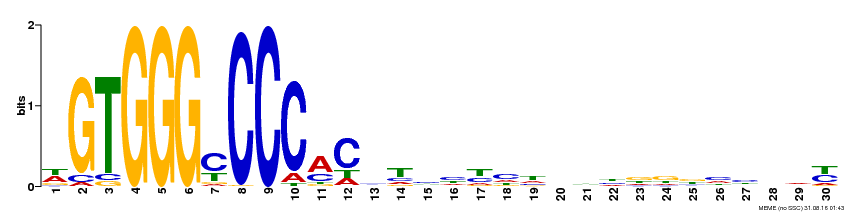
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00215 | DAP | Transfer from AT1G67260 | Download |
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_012077933.2 | 0.0 | transcription factor CYCLOIDEA | ||||
| TrEMBL | A0A067KM92 | 0.0 | A0A067KM92_JATCU; Uncharacterized protein | ||||
| STRING | cassava4.1_022722m | 2e-90 | (Manihot esculenta) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF1738 | 32 | 94 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G67260.2 | 3e-29 | TCP family protein | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



