 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | KDD72353.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Chlorophyta; Trebouxiophyceae; Chlorellales
|
||||||||
| Family | MYB | ||||||||
| Protein Properties | Length: 250aa MW: 28687.6 Da PI: 9.9201 | ||||||||
| Description | MYB family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 42.2 | 1.9e-13 | 8 | 52 | 3 | 48 |
SS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHHT CS
Myb_DNA-binding 3 rWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqkyl 48
W Ede+l+ av ++G ++W++I++ + ++++kqck rw+ +l
KDD72353.1 8 IWKNTEDEILKAAVMKYGLNQWSRISSLLV-RKSAKQCKARWYEWL 52
5999**************************.************986 PP
| |||||||
| 2 | Myb_DNA-binding | 34.1 | 6.5e-11 | 59 | 101 | 2 | 47 |
SSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHH CS
Myb_DNA-binding 2 grWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqky 47
WT eEde+l+++ k++++ W+tIa +g Rt+ qc +r+ +
KDD72353.1 59 TEWTREEDEKLLHLAKLMPTQ-WRTIAPVVG--RTPTQCLERYERL 101
68*****************99.********8..*********9876 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51294 | 21.206 | 1 | 56 | IPR017930 | Myb domain |
| SMART | SM00717 | 8.7E-14 | 5 | 54 | IPR001005 | SANT/Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 4.9E-19 | 8 | 60 | IPR009057 | Homeodomain-like |
| CDD | cd00167 | 1.32E-11 | 9 | 52 | No hit | No description |
| Pfam | PF13921 | 8.4E-14 | 9 | 69 | No hit | No description |
| SuperFamily | SSF46689 | 7.19E-21 | 32 | 104 | IPR009057 | Homeodomain-like |
| CDD | cd11659 | 1.34E-30 | 54 | 106 | No hit | No description |
| PROSITE profile | PS51294 | 15.911 | 57 | 106 | IPR017930 | Myb domain |
| SMART | SM00717 | 1.7E-12 | 57 | 104 | IPR001005 | SANT/Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 4.5E-14 | 61 | 103 | IPR009057 | Homeodomain-like |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009870 | Biological Process | defense response signaling pathway, resistance gene-dependent | ||||
| GO:0010204 | Biological Process | defense response signaling pathway, resistance gene-independent | ||||
| GO:0042742 | Biological Process | defense response to bacterium | ||||
| GO:0050832 | Biological Process | defense response to fungus | ||||
| GO:0009507 | Cellular Component | chloroplast | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 250 aa Download sequence Send to blast |
MFLIKGGIWK NTEDEILKAA VMKYGLNQWS RISSLLVRKS AKQCKARWYE WLDPAIKKTE 60 WTREEDEKLL HLAKLMPTQW RTIAPVVGRT PTQCLERYER LLDAATGEDG GAGSSRDDPR 120 RLRPGEIDPN PESKPARPDP IDMDEDEKEM LSEARARLAN TKGKKAKRKA RERALDEARR 180 LATLQKKREL KAAGIETRDR GPRRGAIDYG TEVPFERRPA AGFHDVEEER ERAQSLRAEF 240 RPATVEQLEG |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 5mqf_L | 1e-115 | 2 | 250 | 4 | 249 | Cell division cycle 5-like protein |
| 5xjc_L | 1e-115 | 2 | 250 | 4 | 249 | Cell division cycle 5-like protein |
| 5yzg_L | 1e-115 | 2 | 250 | 4 | 249 | Cell division cycle 5-like protein |
| 5z56_L | 1e-115 | 2 | 250 | 4 | 249 | Cell division cycle 5-like protein |
| 5z57_L | 1e-115 | 2 | 250 | 4 | 249 | Cell division cycle 5-like protein |
| 5z58_L | 1e-115 | 2 | 250 | 4 | 249 | Cell division cycle 5-like protein |
| 6ff4_L | 1e-115 | 2 | 250 | 4 | 249 | Cell division cycle 5-like protein |
| 6ff7_L | 1e-115 | 2 | 250 | 4 | 249 | Cell division cycle 5-like protein |
| 6icz_L | 1e-115 | 2 | 250 | 4 | 249 | Cell division cycle 5-like protein |
| 6id0_L | 1e-115 | 2 | 250 | 4 | 249 | Cell division cycle 5-like protein |
| 6id1_L | 1e-115 | 2 | 250 | 4 | 249 | Cell division cycle 5-like protein |
| 6qdv_O | 1e-115 | 2 | 250 | 4 | 249 | Cell division cycle 5-like protein |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Component of the MAC complex that probably regulates defense responses through transcriptional control and thereby is essential for plant innate immunity. Possesses a sequence specific DNA sequence 'CTCAGCG' binding activity. Involved in mRNA splicing and cell cycle control. May also play a role in the response to DNA damage. {ECO:0000250|UniProtKB:Q99459, ECO:0000269|PubMed:17298883, ECO:0000269|PubMed:17575050, ECO:0000269|PubMed:8917598}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00028 | SELEX | Transfer from AT1G09770 | Download |
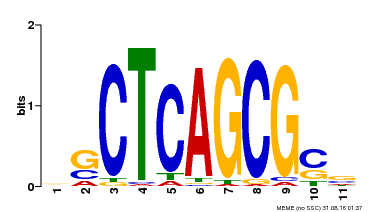
| |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_005648666.1 | 1e-136 | hypothetical protein COCSUDRAFT_65754 | ||||
| Swissprot | P92948 | 1e-122 | CDC5L_ARATH; Cell division cycle 5-like protein | ||||
| TrEMBL | A0A059LDG0 | 1e-179 | A0A059LDG0_9CHLO; Uncharacterized protein (Fragment) | ||||
| STRING | XP_005648666.1 | 1e-135 | (Coccomyxa subellipsoidea) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Chlorophytae | OGCP1040 | 16 | 17 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G09770.1 | 1e-117 | cell division cycle 5 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



