 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | KFK29747.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Arabideae; Arabis
|
||||||||
| Family | B3 | ||||||||
| Protein Properties | Length: 511aa MW: 58029.5 Da PI: 6.9381 | ||||||||
| Description | B3 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | B3 | 34.1 | 4.8e-11 | 26 | 91 | 20 | 99 |
HHH.HTT---..--SEEEEEETTS-EEEEEE..EEETTEEEE-TTHHHHHHHHT--TT-EEEEEE-SSSEE..EEEEE-S CS
B3 20 kkfaeehggkkeesktltledesgrsWevkliyrkksgryvltkGWkeFvkangLkegDfvvFkldgrsefelvvkvfrk 99
+f++++ ++ l +d+s r W+v+l ++ + l++GW+ F+ +n+ ++gD++vF++dg+++ +v+v ++
KFK29747.1 26 DEFLRNYTKV------LLRTDTSERFWKVYL--DGDR----LAGGWEVFAGDNKFRDGDVLVFRHDGDEN--IHVSVSSR 91
5555555422......33367999*******..9998....***********************887554..58888776 PP
| |||||||
| 2 | B3 | 64.6 | 1.5e-20 | 152 | 248 | 1 | 99 |
EEEE-..-HHHHTT-EE--HHH.HTT---..--SEEEEEETTS-EEEEEE..EEETTEEEE-TTHHHHHHHHT--TT-EEEEEE-SSSEE..EEEEE-S CS
B3 1 ffkvltpsdvlksgrlvlpkkfaeehggkkeesktltledesgrsWevkliyrkksgryvltkGWkeFvkangLkegDfvvFkldgrsefelvvkvfrk 99
f+++ ++ ++l ++rl l+++ + + g+ ++ ++ l +++gr+W++ + + +sg +++++GW++F++ ngLk+gD + Fkl++ +++++v+ ++++
KFK29747.1 152 FVTAHVTRYSLHKDRLDLSRNLTVSLGEH-NKAHEIDLVNKQGRRWTLVIAKNCSSGVFYIRRGWSNFCSTNGLKQGDLCKFKLVQ-NGEKPVLWLCSH 248
7888999****************999544.45669***************66666666**************************99.677789999886 PP
| |||||||
| 3 | B3 | 64.3 | 1.9e-20 | 294 | 391 | 1 | 99 |
EEEE-..-HHHHTT-EE--HHH.HTT---..--SEEEEEETTS-EEEEEE....EEETTEEEE-TTHHHHHHHHT--TT-EEEEEE-SSSEE..EEEEE CS
B3 1 ffkvltpsdvlksgrlvlpkkfaeehggkkeesktltledesgrsWevkliy..rkksgryvltkGWkeFvkangLkegDfvvFkldgrsefelvvkvf 97
f+ ++ +++ +k+g+l+l+ f++e+g+ k++ + tl++++gr+W+ + + + +++++++GW+e++kang+k++D++v++l++ +++++v+k++
KFK29747.1 294 FLITEFTPNRFKNGQLYLSTGFTTENGITKPG--ETTLQNKDGRTWSSDIHMtgVEGNRWFYMRRGWREMCKANGVKVNDSFVLELVW-EDANPVFKFH 389
66678899******************999766..79999***********8776666677****************************.89999*9998 PP
-S CS
B3 98 rk 99
+k
KFK29747.1 390 SK 391
76 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SMART | SM01019 | 2.2E-12 | 10 | 92 | IPR003340 | B3 DNA binding domain |
| CDD | cd10017 | 1.96E-6 | 27 | 88 | No hit | No description |
| Gene3D | G3DSA:2.40.330.10 | 1.5E-10 | 31 | 90 | IPR015300 | DNA-binding pseudobarrel domain |
| SuperFamily | SSF101936 | 9.42E-10 | 33 | 90 | IPR015300 | DNA-binding pseudobarrel domain |
| PROSITE profile | PS50863 | 10.08 | 34 | 92 | IPR003340 | B3 DNA binding domain |
| Gene3D | G3DSA:2.40.330.10 | 1.0E-17 | 146 | 247 | IPR015300 | DNA-binding pseudobarrel domain |
| SuperFamily | SSF101936 | 8.44E-18 | 151 | 250 | IPR015300 | DNA-binding pseudobarrel domain |
| SMART | SM01019 | 2.0E-26 | 152 | 249 | IPR003340 | B3 DNA binding domain |
| PROSITE profile | PS50863 | 14.311 | 152 | 248 | IPR003340 | B3 DNA binding domain |
| Pfam | PF02362 | 8.3E-19 | 152 | 248 | IPR003340 | B3 DNA binding domain |
| CDD | cd10017 | 3.76E-16 | 162 | 247 | No hit | No description |
| Gene3D | G3DSA:2.40.330.10 | 2.3E-17 | 289 | 391 | IPR015300 | DNA-binding pseudobarrel domain |
| SuperFamily | SSF101936 | 2.16E-16 | 292 | 390 | IPR015300 | DNA-binding pseudobarrel domain |
| SMART | SM01019 | 1.7E-19 | 294 | 392 | IPR003340 | B3 DNA binding domain |
| PROSITE profile | PS50863 | 11.787 | 294 | 392 | IPR003340 | B3 DNA binding domain |
| Pfam | PF02362 | 2.2E-21 | 294 | 391 | IPR003340 | B3 DNA binding domain |
| CDD | cd10017 | 7.45E-14 | 307 | 390 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009908 | Biological Process | flower development | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 511 aa Download sequence Send to blast |
MANPPRSSLF QQRFLTGNNP VLKLDDEFLR NYTKVLLRTD TSERFWKVYL DGDRLAGGWE 60 VFAGDNKFRD GDVLVFRHDG DENIHVSVSS RSNSGDIEHA SPCNVESDSE YEYEYDDDDG 120 EDEDDEDDDD GLGNISVRKN KKLEADSSSG RFVTAHVTRY SLHKDRLDLS RNLTVSLGEH 180 NKAHEIDLVN KQGRRWTLVI AKNCSSGVFY IRRGWSNFCS TNGLKQGDLC KFKLVQNGEK 240 PVLWLCSHES GNNHEEEECT EVDAVKNCSS GRGKAKNMSN DVSKGKNMKT PSSFLITEFT 300 PNRFKNGQLY LSTGFTTENG ITKPGETTLQ NKDGRTWSSD IHMTGVEGNR WFYMRRGWRE 360 MCKANGVKVN DSFVLELVWE DANPVFKFHS KIENNGSKGK GNRRTRKKRA CETDVTVEKT 420 PTIGIEGRTR VSYRDTTTNS RKLQRTEPKS CSISDQVTNV KRSIVDTLNT VKQFRSELET 480 REQNLEASLL EIDALGERIL GISQILNHKL V |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | May play a role in flower development. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00602 | PBM | Transfer from AT4G31610 | Download |
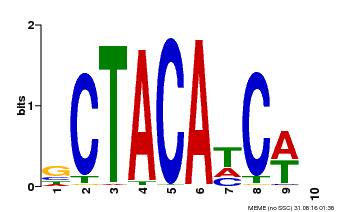
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | KFK29747.1 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_006412578.1 | 0.0 | B3 domain-containing protein REM1 | ||||
| Swissprot | Q84J39 | 0.0 | REM1_ARATH; B3 domain-containing protein REM1 | ||||
| TrEMBL | A0A087GIP5 | 0.0 | A0A087GIP5_ARAAL; Uncharacterized protein | ||||
| STRING | A0A087GIP5 | 0.0 | (Arabis alpina) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM9950 | 17 | 36 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G31610.1 | 1e-180 | B3 family protein | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



