 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | KFK34138.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Arabideae; Arabis
|
||||||||
| Family | C2H2 | ||||||||
| Protein Properties | Length: 1378aa MW: 154855 Da PI: 8.5672 | ||||||||
| Description | C2H2 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | zf-C2H2 | 12.5 | 0.00045 | 1261 | 1286 | 1 | 23 |
EEET..TTTEEESSHHHHHHHHHH.T CS
zf-C2H2 1 ykCp..dCgksFsrksnLkrHirt.H 23
++C C++sF++ +L +H r+ +
KFK34138.1 1261 FQCDleGCTMSFNSEKELAVHKRNiC 1286
89********************9877 PP
| |||||||
| 2 | zf-C2H2 | 11.8 | 0.00077 | 1286 | 1308 | 3 | 23 |
ET..TTTEEESSHHHHHHHHHHT CS
zf-C2H2 3 Cp..dCgksFsrksnLkrHirtH 23
Cp Cgk F ++ +L +H r+H
KFK34138.1 1286 CPvkGCGKNFFSHKYLLQHRRVH 1308
9999*****************99 PP
| |||||||
| 3 | zf-C2H2 | 12 | 0.00067 | 1344 | 1370 | 1 | 23 |
EEET..TTTEEESSHHHHHHHHHH..T CS
zf-C2H2 1 ykCp..dCgksFsrksnLkrHirt..H 23
y+C Cg++F+ s++ rH r+ H
KFK34138.1 1344 YVCAepGCGQTFRFVSDFSRHKRKtgH 1370
899999****************99666 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SMART | SM00545 | 1.0E-15 | 19 | 60 | IPR003349 | JmjN domain |
| PROSITE profile | PS51183 | 13.786 | 20 | 61 | IPR003349 | JmjN domain |
| Pfam | PF02375 | 7.8E-14 | 21 | 54 | IPR003349 | JmjN domain |
| SMART | SM00558 | 2.2E-51 | 200 | 369 | IPR003347 | JmjC domain |
| PROSITE profile | PS51184 | 33.823 | 203 | 369 | IPR003347 | JmjC domain |
| SuperFamily | SSF51197 | 4.39E-27 | 218 | 387 | No hit | No description |
| Pfam | PF02373 | 1.7E-37 | 234 | 352 | IPR003347 | JmjC domain |
| SMART | SM00355 | 12 | 1261 | 1283 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE profile | PS50157 | 12.902 | 1284 | 1313 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 0.02 | 1284 | 1308 | IPR015880 | Zinc finger, C2H2-like |
| Gene3D | G3DSA:3.30.160.60 | 9.2E-6 | 1286 | 1312 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| PROSITE pattern | PS00028 | 0 | 1286 | 1308 | IPR007087 | Zinc finger, C2H2 |
| SuperFamily | SSF57667 | 6.97E-10 | 1300 | 1342 | No hit | No description |
| Gene3D | G3DSA:3.30.160.60 | 9.8E-9 | 1313 | 1337 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| PROSITE profile | PS50157 | 10.928 | 1314 | 1343 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 0.0015 | 1314 | 1338 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE pattern | PS00028 | 0 | 1316 | 1338 | IPR007087 | Zinc finger, C2H2 |
| SuperFamily | SSF57667 | 3.04E-8 | 1332 | 1366 | No hit | No description |
| Gene3D | G3DSA:3.30.160.60 | 1.8E-9 | 1338 | 1367 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| PROSITE profile | PS50157 | 11.032 | 1344 | 1375 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 0.62 | 1344 | 1370 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE pattern | PS00028 | 0 | 1346 | 1370 | IPR007087 | Zinc finger, C2H2 |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009741 | Biological Process | response to brassinosteroid | ||||
| GO:0009826 | Biological Process | unidimensional cell growth | ||||
| GO:0010228 | Biological Process | vegetative to reproductive phase transition of meristem | ||||
| GO:0033169 | Biological Process | histone H3-K9 demethylation | ||||
| GO:0035067 | Biological Process | negative regulation of histone acetylation | ||||
| GO:0048366 | Biological Process | leaf development | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003676 | Molecular Function | nucleic acid binding | ||||
| GO:0046872 | Molecular Function | metal ion binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 1378 aa Download sequence Send to blast |
MAVSEQSQDV FPWLKSLPVA PEFRPTLAEF QDPIAYIFKI EEEASRYGIC KILPPVPPPS 60 KKTAISNLNR SLAARAAARV RDGGGSAFDY DGGPTFATRQ QQIGFCPRKQ RPVQRPVWQS 120 GEHYSFGEFE YKAKTFEKNY LKKCSKKSQL SALEMETLYW KATVDKPFSV EYANDMPGSA 180 FIPLSLAAAR RREYGGDGGT VGETAWNMRA MARAEGSLLK FMKEEIPGVT SPMIYIAMMF 240 SWFAWHVEDH DLHSLNYLHM GASKTWYGVP KDAAVAFEEV VRVHGYGGEL NPLVTFSTLG 300 EKTTVMSPEV FVKAGIPCCR LVQNPGDFVV TFPRAYHSGF SHGFNCGEAS NIATPEWLRM 360 AKDAAIRRAA ISYRPIVSHL QLLYDFALAL GSRVPTSIHT KPRSSRLKDK KRSEGEKLTK 420 ELFVQNIIHN NELLHSLGKG SPIALLPQSS SDKSVCSDQR KSLHLYXXXX MVSLSNGVKD 480 TVSVKEKFTS LCERNRNHLE KETQGTLTDA ERRKNVRLSD QRLFSCVTCG ILSFDCVAIV 540 QPKEAAARYL MSADCSFFND WTVASGSANL GQVARSLHPH CSFFNDWTVA SGSANLGQVA 600 RSLHPQSMEK HDADYFYDVP IEIMDQRTSS TSLTKAQKDN DALGLLASAY GDSSDSEEED 660 HKGIDIPISE GAFKASSVDT DGKEEARDGR SSDFNSQRLS CGKGKEVEVS HATSTCSTLS 720 CTSKQNKLSK GDNTSLVEIA LPFVPRSDDD SSRLHVFCLE HAAEVEEQLR PIGGIHIMLL 780 CHPEYPRIEA EAKIVAEELG VNQEWNDTEF RNVTQEDEET IQAALDNVGT KAGNSDWAVK 840 LGINLSYSAI LSRSPLYSKQ MPYNSIIYNA FGRSSPATSS PSKLEISGKR SSRQRKYVVG 900 KWCGKVWMSH QVHPFLLQQD LEGEESDRSH HLRVALDEDV TGKRLFPGNN SRDATTMFGR 960 KYSRKRKMRA KAVPRKKLTS FKREDGVSDD TSEDHSYKQQ WRASDNEEES YFEIGNTGSG 1020 DSSNHMSDHQ QLKGSRRHKG LKEFESGDEA SDRSLGEEYA VRDYAVSESS MENSFELYRE 1080 KQSRYDHEDD DLYRYPRGIP RNKRAKVFRN PVSYDSEENG AYQQRRRVST SKQARRMGGE 1140 YDSAENSLEE QNFSVTGKRQ TRSTAKRKAK IEAVQSPSDT EGCGLQGFAS GKKNRELDSY 1200 MEGPSTRLRV RNLKPSRGSK VTKPKKTGRK GISIATFSRA ASEEELEEDE GENEEEERTA 1260 FQCDLEGCTM SFNSEKELAV HKRNICPVKG CGKNFFSHKY LLQHRRVHSD DRPLKCPWKG 1320 CKMTFKWAWS RTEHIRVHTG ARPYVCAEPG CGQTFRFVSD FSRHKRKTGH SVKKTKKR |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 6a57_A | 4e-74 | 1260 | 1378 | 22 | 140 | Lysine-specific demethylase REF6 |
| 6a58_A | 4e-74 | 1260 | 1378 | 22 | 140 | Lysine-specific demethylase REF6 |
| 6a59_A | 4e-74 | 1260 | 1378 | 22 | 140 | Lysine-specific demethylase REF6 |
| 6ip0_A | 4e-73 | 9 | 465 | 4 | 421 | Transcription factor jumonji (Jmj) family protein |
| 6ip4_A | 4e-73 | 9 | 465 | 4 | 421 | Arabidopsis JMJ13 |
| Search in ModeBase | ||||||
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 962 | 967 | SRKRKM |
| 2 | 964 | 976 | KRKMRAKAVPRKK |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Histone demethylase that demethylates 'Lys-27' (H3K27me) of histone H3. Demethylates both tri- (H3K27me3) and di-methylated (H3K27me2) H3K27me (PubMed:21642989, PubMed:27111035). Demethylates also H3K4me3/2 and H3K36me3/2 in an in vitro assay (PubMed:20711170). Involved in the transcriptional regulation of hundreds of genes regulating developmental patterning and responses to various stimuli (PubMed:18467490). Binds DNA via its four zinc fingers in a sequence-specific manner, 5'-CTCTGYTY-3', to promote the demethylation of H3K27me3 and the regulation of organ boundary formation (PubMed:27111034, PubMed:27111035). Involved in the regulation of flowering time by repressing FLOWERING LOCUS C (FLC) expression (PubMed:15377760). Interacts with the NF-Y complexe to regulate SOC1 (PubMed:25105952). Mediates the recruitment of BRM to its target loci (PubMed:27111034). {ECO:0000269|PubMed:15377760, ECO:0000269|PubMed:18467490, ECO:0000269|PubMed:20711170, ECO:0000269|PubMed:21642989, ECO:0000269|PubMed:25105952, ECO:0000269|PubMed:27111034, ECO:0000269|PubMed:27111035}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00608 | ChIP-seq | Transfer from AT3G48430 | Download |
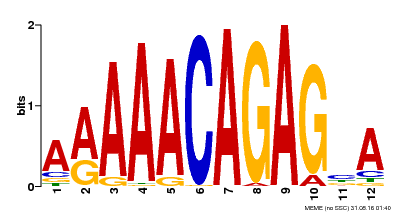
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | KFK34138.1 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AY664499 | 0.0 | AY664499.1 Arabidopsis thaliana relative of early flowering 6 (REF6) mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_006404261.1 | 0.0 | lysine-specific demethylase REF6 | ||||
| Swissprot | Q9STM3 | 0.0 | REF6_ARATH; Lysine-specific demethylase REF6 | ||||
| TrEMBL | A0A087GW86 | 0.0 | A0A087GW86_ARAAL; Uncharacterized protein | ||||
| STRING | A0A087GW86 | 0.0 | (Arabis alpina) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM5259 | 28 | 43 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G48430.1 | 0.0 | relative of early flowering 6 | ||||



