 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | KFK37142.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Arabideae; Arabis
|
||||||||
| Family | NAC | ||||||||
| Protein Properties | Length: 250aa MW: 29170.4 Da PI: 7.062 | ||||||||
| Description | NAC family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | NAM | 88.7 | 1e-27 | 16 | 93 | 2 | 80 |
NAM 2 ppGfrFhPtdeelvveyLkkkvegkkleleevikevdiykvePwdLpkkvkaeekewyfFskrdkkyatgkrknratks 80
pGfrFhPtdeel+ +yL++kve+k+ ++ e ik++diyk++PwdLp+ + +ekewyfF+ r +ky+++ r+nr+t s
KFK37142.1 16 LPGFRFHPTDEELLGFYLRRKVENKPSRI-ELIKQIDIYKYDPWDLPRVSSVGEKEWYFFCMRGRKYRNSVRPNRVTGS 93
69************************999.89***************7777799*********************9875 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF101941 | 9.29E-39 | 15 | 132 | IPR003441 | NAC domain |
| PROSITE profile | PS51005 | 24.512 | 15 | 180 | IPR003441 | NAC domain |
| Pfam | PF02365 | 5.3E-13 | 17 | 109 | IPR003441 | NAC domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0005992 | Biological Process | trehalose biosynthetic process | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0006561 | Biological Process | proline biosynthetic process | ||||
| GO:0009718 | Biological Process | anthocyanin-containing compound biosynthetic process | ||||
| GO:0010120 | Biological Process | camalexin biosynthetic process | ||||
| GO:0042538 | Biological Process | hyperosmotic salinity response | ||||
| GO:1900056 | Biological Process | negative regulation of leaf senescence | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 250 aa Download sequence Send to blast |
MSGESKHHED DDEATLPGFR FHPTDEELLG FYLRRKVENK PSRIELIKQI DIYKYDPWDL 60 PRVSSVGEKE WYFFCMRGRK YRNSVRPNRV TGSAGKGTKT DWMMHEFRLP ATTKTDSPVQ 120 QAEVWTLCRI FKRVTLQRNP TIVPPNRKPV MTLTDSCSKT SSLDSDHTSH HHVVDSLYHK 180 LHEPQPPQTQ TQNPYWNQYT VGYNQPSTYT CNDNNFLSSW NLNDEDFIGD SASWDELRSV 240 IDGNNTKPEK |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1ut4_A | 2e-32 | 17 | 133 | 19 | 165 | NO APICAL MERISTEM PROTEIN |
| 1ut4_B | 2e-32 | 17 | 133 | 19 | 165 | NO APICAL MERISTEM PROTEIN |
| 1ut7_A | 2e-32 | 17 | 133 | 19 | 165 | NO APICAL MERISTEM PROTEIN |
| 1ut7_B | 2e-32 | 17 | 133 | 19 | 165 | NO APICAL MERISTEM PROTEIN |
| 4dul_A | 2e-32 | 17 | 133 | 19 | 165 | NAC domain-containing protein 19 |
| 4dul_B | 2e-32 | 17 | 133 | 19 | 165 | NAC domain-containing protein 19 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription factor that binds to the 5'- RRYGCCGT-3' consensus core sequence. Central longevity regulator. Negative regulator of leaf senescence. Modulates cellular H(2)O(2) levels and enhances tolerance to various abiotic stresses through the regulation of DREB2A. {ECO:0000269|PubMed:22345491}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00310 | DAP | Transfer from AT2G43000 | Download |
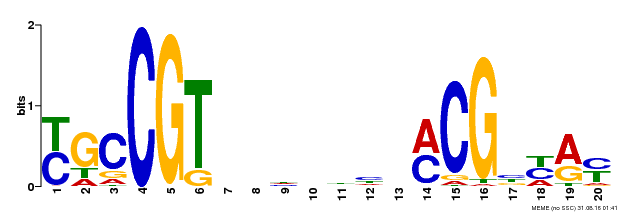
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | KFK37142.1 |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Up-regulated by H(2)O(2), paraquat, ozone, 3-aminotriazole and salt stress. {ECO:0000269|PubMed:22345491}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_009142897.1 | 1e-142 | PREDICTED: transcription factor JUNGBRUNNEN 1-like | ||||
| Swissprot | Q9SK55 | 1e-138 | NAC42_ARATH; Transcription factor JUNGBRUNNEN 1 | ||||
| TrEMBL | A0A087H4U0 | 0.0 | A0A087H4U0_ARAAL; Uncharacterized protein | ||||
| STRING | A0A087H4U0 | 0.0 | (Arabis alpina) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM306 | 28 | 200 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G43000.1 | 1e-116 | NAC domain containing protein 42 | ||||



