 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | KFK41389.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Arabideae; Arabis
|
||||||||
| Family | HD-ZIP | ||||||||
| Protein Properties | Length: 297aa MW: 33605.3 Da PI: 5.404 | ||||||||
| Description | HD-ZIP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Homeobox | 56 | 6.8e-18 | 88 | 141 | 3 | 56 |
--SS--HHHHHHHHHHHHHSSS--HHHHHHHHHHCTS-HHHHHHHHHHHHHHHH CS
Homeobox 3 kRttftkeqleeLeelFeknrypsaeereeLAkklgLterqVkvWFqNrRakek 56
k+++++ eq+++Le+ Fe +++ e++ +LA+ lgL+ rq+ +WFqNrRa++k
KFK41389.1 88 KKRRLNMEQVKTLEKNFELGNKLEPERKMQLARALGLQPRQIAIWFQNRRARWK 141
456899***********************************************9 PP
| |||||||
| 2 | HD-ZIP_I/II | 130.5 | 6.7e-42 | 87 | 178 | 1 | 92 |
HD-ZIP_I/II 1 ekkrrlskeqvklLEesFeeeekLeperKvelareLglqprqvavWFqnrRARtktkqlEkdyeaLkraydalkeenerLekeveeLreelk 92
ekkrrl+ eqvk+LE++Fe +kLeperK++lar+Lglqprq+a+WFqnrRAR+ktkqlEkdy++Lk+++dalk+en+ L++++++L+ e++
KFK41389.1 87 EKKRRLNMEQVKTLEKNFELGNKLEPERKMQLARALGLQPRQIAIWFQNRRARWKTKQLEKDYDTLKKQFDALKAENDLLQTHNQKLQVEIM 178
69*************************************************************************************98875 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF46689 | 1.58E-19 | 77 | 145 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS50071 | 17.216 | 83 | 143 | IPR001356 | Homeobox domain |
| SMART | SM00389 | 1.8E-17 | 86 | 147 | IPR001356 | Homeobox domain |
| CDD | cd00086 | 3.51E-16 | 88 | 144 | No hit | No description |
| Pfam | PF00046 | 3.0E-15 | 88 | 141 | IPR001356 | Homeobox domain |
| Gene3D | G3DSA:1.10.10.60 | 5.3E-19 | 90 | 150 | IPR009057 | Homeodomain-like |
| PRINTS | PR00031 | 1.2E-5 | 114 | 123 | IPR000047 | Helix-turn-helix motif |
| PROSITE pattern | PS00027 | 0 | 118 | 141 | IPR017970 | Homeobox, conserved site |
| PRINTS | PR00031 | 1.2E-5 | 123 | 139 | IPR000047 | Helix-turn-helix motif |
| Pfam | PF02183 | 6.6E-15 | 143 | 184 | IPR003106 | Leucine zipper, homeobox-associated |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009744 | Biological Process | response to sucrose | ||||
| GO:0048826 | Biological Process | cotyledon morphogenesis | ||||
| GO:0080022 | Biological Process | primary root development | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 297 aa Download sequence Send to blast |
MSCNNGMSFF PSNFMIQTSY EEDNHHPNQS PSLAPLLPSC TSLPQDLHGF ASFLGKRSPV 60 EGCCDLETGN NMNGEEDYSD DGSQMGEKKR RLNMEQVKTL EKNFELGNKL EPERKMQLAR 120 ALGLQPRQIA IWFQNRRARW KTKQLEKDYD TLKKQFDALK AENDLLQTHN QKLQVEIMGL 180 KNREQTESIN LNKETEGSCS NRSDNSSDNL RLDISTAPPS IDSTLTGSHQ PPPQTLGRHF 240 FPPSPAAATT TATTVQFFHN SSSGQSMVKE ENSISNMFCA MDDQSGFWPW LDQQQYN |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 135 | 143 | RRARWKTKQ |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcription factor that may act in the sucrose-signaling pathway. {ECO:0000269|PubMed:11292072}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00225 | DAP | Transfer from AT1G69780 | Download |
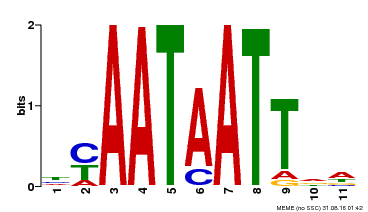
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | KFK41389.1 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AF208044 | 0.0 | AF208044.1 Arabidopsis thaliana homeodomain leucine-zipper protein ATHB13 mRNA, complete cds. | |||
| GenBank | AY057572 | 0.0 | AY057572.1 Arabidopsis thaliana At1g69780/T6C23_2 mRNA, complete cds. | |||
| GenBank | AY133645 | 0.0 | AY133645.1 Arabidopsis thaliana At1g69780/T6C23_2 mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | NP_177136.1 | 0.0 | Homeobox-leucine zipper protein family | ||||
| Swissprot | Q8LC03 | 0.0 | ATB13_ARATH; Homeobox-leucine zipper protein ATHB-13 | ||||
| TrEMBL | A0A087HGY7 | 0.0 | A0A087HGY7_ARAAL; Uncharacterized protein | ||||
| STRING | A0A087HGY7 | 0.0 | (Arabis alpina) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM2633 | 27 | 70 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G69780.1 | 0.0 | HD-ZIP family protein | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



