 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | KFK41622.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Arabideae; Arabis
|
||||||||
| Family | C2H2 | ||||||||
| Protein Properties | Length: 373aa MW: 42580.2 Da PI: 7.8278 | ||||||||
| Description | C2H2 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | zf-C2H2 | 11.2 | 0.0012 | 20 | 42 | 1 | 23 |
EEETTTTEEESSHHHHHHHHHHT CS
zf-C2H2 1 ykCpdCgksFsrksnLkrHirtH 23
y C++Cg s s k++ ++Hi +H
KFK41622.1 20 YLCQYCGISRSKKYLITSHILSH 42
56*******************96 PP
| |||||||
| 2 | zf-C2H2 | 18.7 | 4.8e-06 | 66 | 88 | 1 | 23 |
EEETTTTEEESSHHHHHHHHHHT CS
zf-C2H2 1 ykCpdCgksFsrksnLkrHirtH 23
+ C+ Cg F+ + +Lk+H+ +H
KFK41622.1 66 HNCQECGAEFKKPAHLKQHMLSH 88
67*******************99 PP
| |||||||
| 3 | zf-C2H2 | 15.5 | 4.8e-05 | 94 | 118 | 1 | 23 |
EEET..TTTEEESSHHHHHHHHHHT CS
zf-C2H2 1 ykCp..dCgksFsrksnLkrHirtH 23
++C dC s++rk++L+rH+ tH
KFK41622.1 94 FTCYvdDCAASYRRKDHLNRHLLTH 118
789999*****************99 PP
| |||||||
| 4 | zf-C2H2 | 22 | 4.3e-07 | 175 | 199 | 1 | 23 |
EEET..TTTEEESSHHHHHHHHHHT CS
zf-C2H2 1 ykCp..dCgksFsrksnLkrHirtH 23
++C+ Cgk F+ +s+L++H+ +H
KFK41622.1 175 HVCQevGCGKAFRYPSQLQKHQDSH 199
789999****************998 PP
| |||||||
| 5 | zf-C2H2 | 22.2 | 3.7e-07 | 265 | 290 | 1 | 23 |
EEET..TTTEEESSHHHHHHHHHH.T CS
zf-C2H2 1 ykCp..dCgksFsrksnLkrHirt.H 23
+kC+ C+ +Fs +snL++H++ H
KFK41622.1 265 FKCEveGCSSTFSKPSNLQKHMKAvH 290
89********************9888 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SMART | SM00355 | 25 | 20 | 42 | IPR015880 | Zinc finger, C2H2-like |
| SMART | SM00355 | 0.0069 | 66 | 88 | IPR015880 | Zinc finger, C2H2-like |
| Gene3D | G3DSA:3.30.160.60 | 1.5E-4 | 66 | 84 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| PROSITE profile | PS50157 | 13.339 | 66 | 93 | IPR007087 | Zinc finger, C2H2 |
| PROSITE pattern | PS00028 | 0 | 68 | 88 | IPR007087 | Zinc finger, C2H2 |
| SuperFamily | SSF57667 | 1.59E-10 | 78 | 120 | No hit | No description |
| Gene3D | G3DSA:3.30.160.60 | 8.0E-13 | 85 | 120 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| SMART | SM00355 | 0.11 | 94 | 118 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE profile | PS50157 | 9.91 | 94 | 123 | IPR007087 | Zinc finger, C2H2 |
| PROSITE pattern | PS00028 | 0 | 96 | 118 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 0.24 | 123 | 148 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE profile | PS50157 | 8.808 | 123 | 153 | IPR007087 | Zinc finger, C2H2 |
| PROSITE pattern | PS00028 | 0 | 125 | 148 | IPR007087 | Zinc finger, C2H2 |
| SuperFamily | SSF57667 | 4.61E-7 | 171 | 201 | No hit | No description |
| Gene3D | G3DSA:3.30.160.60 | 6.2E-9 | 171 | 199 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| SMART | SM00355 | 0.0022 | 175 | 199 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE profile | PS50157 | 10.949 | 175 | 204 | IPR007087 | Zinc finger, C2H2 |
| PROSITE pattern | PS00028 | 0 | 177 | 199 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 4.8 | 207 | 232 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE pattern | PS00028 | 0 | 209 | 232 | IPR007087 | Zinc finger, C2H2 |
| Gene3D | G3DSA:3.30.160.60 | 7.5E-4 | 209 | 230 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| SMART | SM00355 | 25 | 235 | 256 | IPR015880 | Zinc finger, C2H2-like |
| SuperFamily | SSF57667 | 7.96E-10 | 246 | 303 | No hit | No description |
| SMART | SM00355 | 8.9E-5 | 265 | 290 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE profile | PS50157 | 13.027 | 265 | 295 | IPR007087 | Zinc finger, C2H2 |
| Gene3D | G3DSA:3.30.160.60 | 9.9E-9 | 265 | 290 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| PROSITE pattern | PS00028 | 0 | 267 | 290 | IPR007087 | Zinc finger, C2H2 |
| Gene3D | G3DSA:3.30.160.60 | 1.4E-4 | 291 | 317 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| SMART | SM00355 | 5.3 | 296 | 322 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE pattern | PS00028 | 0 | 298 | 322 | IPR007087 | Zinc finger, C2H2 |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0005730 | Cellular Component | nucleolus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0008097 | Molecular Function | 5S rRNA binding | ||||
| GO:0046872 | Molecular Function | metal ion binding | ||||
| GO:0080084 | Molecular Function | 5S rDNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 373 aa Download sequence Send to blast |
MGEESNIDMD ASLIKDIRKY LCQYCGISRS KKYLITSHIL SHHKMEMEEE RDDEACEVEE 60 EALGKHNCQE CGAEFKKPAH LKQHMLSHSI ERPFTCYVDD CAASYRRKDH LNRHLLTHKG 120 KLFKCPMENC KSEFSVQGNV GRHIKEMHKN GDGSKDDIGN GDSHPTEGSI GQKPHVCQEV 180 GCGKAFRYPS QLQKHQDSHV KLDSVEAFCS EPGCMKYFTN DECLKAHIRS CHQHVNCEIC 240 GSKHLKKNIK RHLRTHDEDS APGVFKCEVE GCSSTFSKPS NLQKHMKAVH EDIRPFICGF 300 PGCGMRFAYK HVRNNHENSG SHVYTCGDFV ATDENFTSRP RGGLKRKQVT AEMLIRKRVM 360 PPQFESQEEH KTC |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 5v3m_C | 2e-17 | 5 | 325 | 19 | 309 | Zinc finger protein 568 |
| 5wjq_D | 1e-17 | 20 | 325 | 9 | 284 | Zinc finger protein 568 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Essential protein (PubMed:22353599). Isoform 1 is a transcription activator the binds both 5S rDNA and 5S rRNA and stimulates the transcription of 5S rRNA gene (PubMed:12711688, PubMed:22353599). Isoform 1 regulates 5S rRNA levels during development (PubMed:22353599). {ECO:0000269|PubMed:12711688, ECO:0000269|PubMed:22353599}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00229 | DAP | Transfer from AT1G72050 | Download |
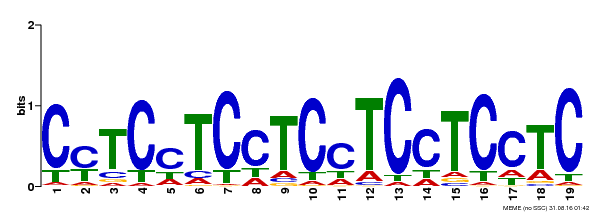
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | KFK41622.1 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010415924.1 | 0.0 | PREDICTED: transcription factor IIIA | ||||
| Swissprot | Q84MZ4 | 0.0 | TF3A_ARATH; Transcription factor IIIA | ||||
| TrEMBL | A0A087HHM0 | 0.0 | A0A087HHM0_ARAAL; Uncharacterized protein | ||||
| STRING | A0A087HHL9 | 0.0 | (Arabis alpina) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G72050.2 | 0.0 | transcription factor IIIA | ||||



