 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | KFK41931.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Arabideae; Arabis
|
||||||||
| Family | ERF | ||||||||
| Protein Properties | Length: 205aa MW: 23154.6 Da PI: 4.2581 | ||||||||
| Description | ERF family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | AP2 | 59.2 | 1e-18 | 22 | 72 | 1 | 55 |
AP2 1 sgykGVrwdkkrgrWvAeIrdpsengkrkrfslgkfgtaeeAakaaiaarkkleg 55
s+ykGVr +k +g+Wv+eIr p++ r r++lg++ t+e+Aa+a +aa +l+g
KFK41931.1 22 SKYKGVRKRK-WGKWVSEIRLPNS---RERIWLGSYETPEKAARAFDAAQFCLRG 72
89*****999.**********933...5*************************98 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF54171 | 2.22E-20 | 22 | 81 | IPR016177 | DNA-binding domain |
| CDD | cd00018 | 2.85E-28 | 22 | 81 | No hit | No description |
| Pfam | PF00847 | 6.0E-12 | 22 | 72 | IPR001471 | AP2/ERF domain |
| PROSITE profile | PS51032 | 21.878 | 23 | 80 | IPR001471 | AP2/ERF domain |
| SMART | SM00380 | 6.0E-29 | 23 | 86 | IPR001471 | AP2/ERF domain |
| Gene3D | G3DSA:3.30.730.10 | 8.7E-28 | 23 | 81 | IPR001471 | AP2/ERF domain |
| PRINTS | PR00367 | 1.7E-9 | 24 | 35 | IPR001471 | AP2/ERF domain |
| PRINTS | PR00367 | 1.7E-9 | 46 | 62 | IPR001471 | AP2/ERF domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0001944 | Biological Process | vasculature development | ||||
| GO:0002213 | Biological Process | defense response to insect | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009611 | Biological Process | response to wounding | ||||
| GO:0051301 | Biological Process | cell division | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 205 aa Download sequence Send to blast |
MAKKAMKKNE EEEEKRNNSL QSKYKGVRKR KWGKWVSEIR LPNSRERIWL GSYETPEKAA 60 RAFDAAQFCL RGGDGNYNFP DRPPFISGGK SLTPSAIQEA AARFANTEEE SVVVVVKGEE 120 EEESGLPGSE IRPESPSPSF VSEVATSTLD CDWSLLDMIP MDFGTFPEFD DFSDVFSGDR 180 FTENLSVGDY GEENIVDDSF FLWDF |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 5wx9_A | 2e-13 | 21 | 88 | 12 | 80 | Ethylene-responsive transcription factor ERF096 |
| Search in ModeBase | ||||||
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 24 | 30 | KGVRKRK |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probably acts as a transcriptional activator. Binds to the GCC-box pathogenesis-related promoter element. May be involved in the regulation of gene expression by stress factors and by components of stress signal transduction pathways (By similarity). {ECO:0000250}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00601 | PBM | Transfer from AT1G74930 | Download |
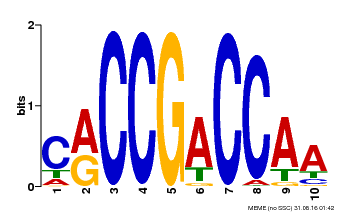
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | KFK41931.1 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AF332422 | 4e-41 | AF332422.1 Arabidopsis thaliana clone C00087 putative AP2 domain transcription factor (At1g19210) mRNA, partial cds. | |||
| GenBank | AY008390 | 4e-41 | AY008390.1 Arabidopsis thaliana ARF11/IAA22 gene, complete cds. | |||
| GenBank | BT006191 | 4e-41 | BT006191.1 Arabidopsis thaliana clone C00087 (K) putative AP2 domain transcription factor (At1g19210) mRNA, complete cds. | |||
| GenBank | CP002684 | 4e-41 | CP002684.1 Arabidopsis thaliana chromosome 1 sequence. | |||
| GenBank | T29M8 | 4e-41 | AC069143.3 Sequence of BAC T29M8 from Arabidopsis thaliana chromosome 1, complete sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010471592.1 | 7e-99 | PREDICTED: ethylene-responsive transcription factor ERF018-like | ||||
| Swissprot | Q9S7L5 | 1e-90 | ERF18_ARATH; Ethylene-responsive transcription factor ERF018 | ||||
| TrEMBL | A0A087HIH9 | 1e-147 | A0A087HIH9_ARAAL; Uncharacterized protein | ||||
| STRING | A0A087HIH9 | 1e-148 | (Arabis alpina) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM1614 | 28 | 90 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G74930.1 | 3e-72 | ERF family protein | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



