 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | KFK44212.1 | ||||||||
| Common Name | SPL14 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Arabideae; Arabis
|
||||||||
| Family | SBP | ||||||||
| Protein Properties | Length: 1024aa MW: 113998 Da PI: 8.3549 | ||||||||
| Description | SBP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | SBP | 127.8 | 4.1e-40 | 128 | 205 | 1 | 78 |
--SSTT-----TT--HHHHHTT--HHHHT-S-EEETTEEEEE-TTTSSEEETTT--SS--S-STTTT-------S--- CS
SBP 1 lCqvegCeadlseakeyhrrhkvCevhskapvvlvsgleqrfCqqCsrfhelsefDeekrsCrrrLakhnerrrkkqa 78
+Cqv++C++dls+ k+yhrrhkvCevhska+++lv +++qrfCqqCsrfh l efDe+krsCrrrLa+hn+rrrk+++
KFK44212.1 128 MCQVDNCTEDLSHGKDYHRRHKVCEVHSKATKALVGKQMQRFCQQCSRFHLLAEFDEGKRSCRRRLAGHNRRRRKTTQ 205
6**************************************************************************875 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:4.10.1100.10 | 5.7E-33 | 121 | 190 | IPR004333 | Transcription factor, SBP-box |
| PROSITE profile | PS51141 | 31.71 | 126 | 203 | IPR004333 | Transcription factor, SBP-box |
| SuperFamily | SSF103612 | 1.7E-36 | 127 | 204 | IPR004333 | Transcription factor, SBP-box |
| Pfam | PF03110 | 2.6E-28 | 129 | 202 | IPR004333 | Transcription factor, SBP-box |
| CDD | cd00204 | 8.50E-9 | 789 | 914 | No hit | No description |
| SuperFamily | SSF48403 | 1.07E-7 | 809 | 916 | IPR020683 | Ankyrin repeat-containing domain |
| Gene3D | G3DSA:1.25.40.20 | 2.2E-7 | 818 | 923 | IPR020683 | Ankyrin repeat-containing domain |
| PROSITE profile | PS50297 | 8.879 | 861 | 933 | IPR020683 | Ankyrin repeat-containing domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0042742 | Biological Process | defense response to bacterium | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0005886 | Cellular Component | plasma membrane | ||||
| GO:0016021 | Cellular Component | integral component of membrane | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 1024 aa Download sequence Send to blast |
MEEVGAQVAT PIFIHQSLSP MGRKRDLYYH MSNRLVQTQQ PQRRDEWNST MWDWDSRRFQ 60 AKPVESDVLR LGNKTQFDLN ANNRKEVEEK GLDLNLGSCL NTVEDTKTTT IRPSKRVRSG 120 SPGGNYPMCQ VDNCTEDLSH GKDYHRRHKV CEVHSKATKA LVGKQMQRFC QQCSRFHLLA 180 EFDEGKRSCR RRLAGHNRRR RKTTQPEEIA SGAVVPGNNT SNANMDLMAL LTALACAQGK 240 NEVKPMGSPA VPDREKLLQI LSKINALPLP MDLVSKLNNI GSLARKNMDR PTVNPQNDMN 300 GASPSTMDLL AVLSATLGSS SPDALAMLSQ GGFGNKDNDK TKLSSYDHCA TTNLEKRTFV 360 GGERSSSSNQ SPSQEDSDSR AQDTRSSLSL QLFTSSPEDE SRPTVASSRK YYSSASSNPV 420 EDRSLSSSPV MQELFPLQTS PETMRSKNHK NTSHRTGCLP LELFGGSNRG VANPNFKGFG 480 QQSGYASSGS DYSPPSLNSD AQDRTGKIVF KLLDKDPSQL PGTLRTEIYN WLSNIPSEME 540 SYIRPGCVVL SVYVAMSPAA WEQLERNLMH RIGVLLQDSH SEFWKNARFI VNTGRQLASH 600 KNGKIRCSKS WRTWNSPELI SVSPVAVVAG EETSLVVRGR SLTNDGISFR STHMGSYVSM 660 EVTGAPCRQA VFEELNVNSF KVNNTQPGFL GRCFIEVENG FRGDSFPLII ANASICKELN 720 RLEEEFHPKS NDMTEEQAHR PTSREEVLCF LNELGWLFQK HQTSEPREQS DFSLYRFNFL 780 LVCSVERDYC AVISTVLDML VERNLVNDEL NRDALDMLDE IQLLNRAVKR KSTNMVELLI 840 HYSALGSSKK LVFLPNKTGP GGITPLHLAA CTSGSDDMVD LLTNDPQEIG LSSWNTLCDA 900 TGQTPYSYAA MRNNHTYNSL VARKLADKRN KQVSLNIEIV DQMGLSKRLS LEINKSSSCA 960 SCTTVALKYQ RRVSGSHRLF PTPIIHSMLA VATVCVCVCV FMHAFPIVRQ GSHFSWGGLD 1020 YGSI |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1ul4_A | 2e-30 | 119 | 202 | 3 | 84 | squamosa promoter binding protein-like 4 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Trans-acting factor that binds specifically to the consensus nucleotide sequence 5'-TNCGTACAA-3' (By similarity). May play a role in plant development. {ECO:0000250, ECO:0000269|PubMed:15703061}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00155 | DAP | Transfer from AT1G20980 | Download |
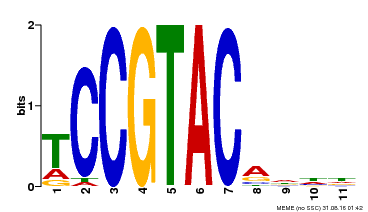
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | KFK44212.1 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AK353554 | 0.0 | AK353554.1 Thellungiella halophila mRNA, complete cds, clone: RTFL01-52-I08. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_006416342.1 | 0.0 | squamosa promoter-binding-like protein 14 | ||||
| Swissprot | Q8RY95 | 0.0 | SPL14_ARATH; Squamosa promoter-binding-like protein 14 | ||||
| TrEMBL | S4WY96 | 0.0 | S4WY96_ARAAL; SQUAMOSA promoter binding protein-like protein 14 | ||||
| STRING | S4WY96 | 0.0 | (Arabis alpina) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM3999 | 25 | 54 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G20980.1 | 0.0 | squamosa promoter binding protein-like 14 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



