 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | KFK44860.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Arabideae; Arabis
|
||||||||
| Family | HD-ZIP | ||||||||
| Protein Properties | Length: 837aa MW: 91971.2 Da PI: 6.4592 | ||||||||
| Description | HD-ZIP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Homeobox | 58.4 | 1.2e-18 | 20 | 78 | 3 | 57 |
--SS--HHHHHHHHHHHHHSSS--HHHHHHHHHHC....TS-HHHHHHHHHHHHHHHHC CS
Homeobox 3 kRttftkeqleeLeelFeknrypsaeereeLAkkl....gLterqVkvWFqNrRakekk 57
k ++t+eq+e+Le++++++++p+ +r++L +++ +++ rq+kvWFqNrR +ek+
KFK44860.1 20 KYVRYTPEQVEALERVYAECPKPTSLRRQQLIRECpilcNIEPRQIKVWFQNRRCREKQ 78
5678*****************************************************97 PP
| |||||||
| 2 | START | 174 | 9.1e-55 | 169 | 377 | 2 | 205 |
HHHHHHHHHHHHHHHC-TT-EEEEEXCCTTEEEEEEESSS.SCEEEEEEEECCSCHHHHHHHHHCCCGGCT-TT-SEEEEEEEECTT..EEEEEEEEXX CS
START 2 laeeaaqelvkkalaeepgWvkssesengdevlqkfeeskvdsgealrasgvvdmvlallveellddkeqWdetlakaetlevissg..galqlmvael 98
+aee+++e++ ka+ ++ Wv+ +++g++++ +++ s+++sg a+ra+g+v +++ v+e+l+d++ W ++++++etl vi +g g+++l ++++
KFK44860.1 169 IAEETLAEFLCKATGTAIDWVQIIGMKPGPDSIGIVAVSRNCSGIAARACGLVGLEPM-KVAEILKDRPSWFRDCRSVETLSVIPTGngGTIELVNTQM 266
7899******************************************************.8888888888****************************** PP
TTXX-SSX.EEEEEEEEEEE.TTS-EEEEEEEEE-TTS--....-TTSEE-EESSEEEEEEEECTCEEEEEEEE-EE--SSXXHHHHHHHHHHHHHHHH CS
START 99 qalsplvp.RdfvfvRyirqlgagdwvivdvSvdseqkppe...sssvvRaellpSgiliepksnghskvtwvehvdlkgrlphwllrslvksglaega 193
+a+++l+p Rdf+++Ry+ +l++g++v++++S++ + p+ s+++vRa++l Sg+li+p+++g+s + +v+hvdl++++++++lr+l++s+ + ++
KFK44860.1 267 YAPTTLAPaRDFWTLRYSTCLEDGSYVVCERSLTFATGGPNtplSPNFVRAKMLSSGFLIRPCDGGGSIINIVDHVDLDASSVPEVLRPLYESSKILAQ 365
**************************************99999******************************************************** PP
HHHHHHTXXXXX CS
START 194 ktwvatlqrqce 205
kt+va+l++ ++
KFK44860.1 366 KTTVAALRHVRQ 377
*******98765 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:1.10.10.60 | 5.0E-19 | 4 | 78 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS50071 | 15.435 | 15 | 79 | IPR001356 | Homeobox domain |
| SMART | SM00389 | 5.9E-16 | 17 | 83 | IPR001356 | Homeobox domain |
| SuperFamily | SSF46689 | 2.1E-16 | 19 | 81 | IPR009057 | Homeodomain-like |
| CDD | cd00086 | 7.09E-17 | 20 | 80 | No hit | No description |
| Pfam | PF00046 | 4.2E-16 | 21 | 78 | IPR001356 | Homeobox domain |
| CDD | cd14686 | 3.20E-6 | 72 | 111 | No hit | No description |
| PROSITE profile | PS50848 | 26.049 | 159 | 387 | IPR002913 | START domain |
| CDD | cd08875 | 7.61E-71 | 163 | 378 | No hit | No description |
| SMART | SM00234 | 2.1E-49 | 168 | 378 | IPR002913 | START domain |
| SuperFamily | SSF55961 | 3.57E-35 | 168 | 379 | No hit | No description |
| Pfam | PF01852 | 1.8E-52 | 169 | 377 | IPR002913 | START domain |
| Gene3D | G3DSA:3.30.530.20 | 1.1E-19 | 205 | 373 | IPR023393 | START-like domain |
| SuperFamily | SSF55961 | 2.61E-5 | 416 | 497 | No hit | No description |
| SuperFamily | SSF55961 | 2.61E-5 | 529 | 604 | No hit | No description |
| Pfam | PF08670 | 6.5E-49 | 693 | 836 | IPR013978 | MEKHLA |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009855 | Biological Process | determination of bilateral symmetry | ||||
| GO:0009880 | Biological Process | embryonic pattern specification | ||||
| GO:0009944 | Biological Process | polarity specification of adaxial/abaxial axis | ||||
| GO:0010072 | Biological Process | primary shoot apical meristem specification | ||||
| GO:0080060 | Biological Process | integument development | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0008289 | Molecular Function | lipid binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 837 aa Download sequence Send to blast |
MMAHSMDDRD SPDKGFDSGK YVRYTPEQVE ALERVYAECP KPTSLRRQQL IRECPILCNI 60 EPRQIKVWFQ NRRCREKQRK ESARLQTVNR KLSAMNKLLM EENDRLQKQV SHLVYENGFM 120 KNRIHTASGT TTDNSCESVV VSGQQRQQQN PTHQHPQRDA NNPAGLLSIA EETLAEFLCK 180 ATGTAIDWVQ IIGMKPGPDS IGIVAVSRNC SGIAARACGL VGLEPMKVAE ILKDRPSWFR 240 DCRSVETLSV IPTGNGGTIE LVNTQMYAPT TLAPARDFWT LRYSTCLEDG SYVVCERSLT 300 FATGGPNTPL SPNFVRAKML SSGFLIRPCD GGGSIINIVD HVDLDASSVP EVLRPLYESS 360 KILAQKTTVA ALRHVRQISQ EASGEVQYTG GRQPAVLRTF SQRLCRGFND AVNGFADDGW 420 TSMSSDGGED ISIMINSSSA KFAGSQYSNS FLPSFGSGVL CAKASMLLQN VPPLVLIRFL 480 REHRAEWADY GVDAYAAASL RATPFTVPCV RSGGFPSNQV IVPLAQTLEH EEDLEVVKLG 540 SHAYSPEDMG LSRDMYLLQV CSGVDENAVG GCAQLVFAPI DESFADDAPL LPSGFRVIPL 600 EHKTTPNDHL SANRTRDLAS SLDGSTKTDS DTNFRLVLTI AFQFTFDNHS RDNVATMARQ 660 YVRNVVGSIQ RVALAITPRP GSMQLPSSPE ALTLVRWISR SYSMHTGANL FGADSDGDTL 720 LKNLWNHTDA ILCCSINTHT SPVFTFANQA GLDMLETTLV ALQDIMLDKT LDGSARKTLC 780 SEFSKITQQG YATLPAGICV SSMGRPVSYE QATVWKVLDN NESNHCLAFI LVNWSFV |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcription factor involved in the determination of adaxial-abaxial polarity in ovule primordium. Specifies adaxial leaf fates. Binds to the DNA sequence 5'-GTAAT[GC]ATTAC-3'. {ECO:0000269|PubMed:11395776, ECO:0000269|PubMed:15598805, ECO:0000269|PubMed:17900520, ECO:0000269|PubMed:9747806}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00170 | DAP | Transfer from AT1G30490 | Download |
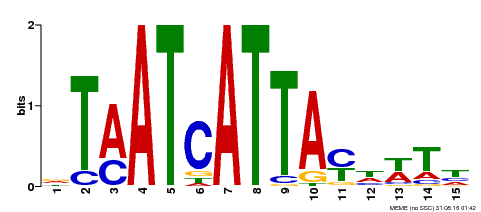
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | KFK44860.1 |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: By auxin. Repressed by miR165 and miR166. {ECO:0000269|PubMed:14999284, ECO:0000269|PubMed:16033795, ECO:0000269|PubMed:17237362}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AK353258 | 0.0 | AK353258.1 Thellungiella halophila mRNA, complete cds, clone: RTFL01-39-H09. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_013662459.1 | 0.0 | homeobox-leucine zipper protein ATHB-9 | ||||
| Swissprot | O04292 | 0.0 | ATBH9_ARATH; Homeobox-leucine zipper protein ATHB-9 | ||||
| TrEMBL | A0A087HRV8 | 0.0 | A0A087HRV8_ARAAL; Uncharacterized protein | ||||
| STRING | A0A087HRV8 | 0.0 | (Arabis alpina) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM4256 | 26 | 53 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G30490.1 | 0.0 | HD-ZIP family protein | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



