 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | KFK45026.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Arabideae; Arabis
|
||||||||
| Family | NAC | ||||||||
| Protein Properties | Length: 342aa MW: 39502 Da PI: 6.051 | ||||||||
| Description | NAC family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | NAM | 164.9 | 2.9e-51 | 16 | 144 | 1 | 128 |
NAM 1 lppGfrFhPtdeelvveyLkkkvegkkleleevikevdiykvePwdLpk..kvka.eekewyfFskrdkkyatgkrknratksgyWkatgkdkevlskk 96
+ppGfrFhPt+eel+++yL+kkv+++k++l +vi+evd++k+ePwd+++ ++ + +++wyfFs++dkky+tg+r+nrat +g+Wkatg+dk ++s
KFK45026.1 16 VPPGFRFHPTEEELLHYYLRKKVNSQKIDL-DVIREVDLNKLEPWDIQEecRIGStPQNDWYFFSHKDKKYPTGTRTNRATVAGFWKATGRDKIIYS-C 112
69****************************.9***************963433332456*************************************9.9 PP
NAM 97 gelvglkktLvfykgrapkgektdWvmheyrl 128
+ +gl+ktLvfykgrap+g+k+dW+mheyrl
KFK45026.1 113 VRRIGLRKTLVFYKGRAPHGQKSDWIMHEYRL 144
999***************************98 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF101941 | 2.09E-56 | 12 | 181 | IPR003441 | NAC domain |
| PROSITE profile | PS51005 | 55.39 | 16 | 181 | IPR003441 | NAC domain |
| Pfam | PF02365 | 6.4E-27 | 17 | 144 | IPR003441 | NAC domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009809 | Biological Process | lignin biosynthetic process | ||||
| GO:0009834 | Biological Process | plant-type secondary cell wall biogenesis | ||||
| GO:0010047 | Biological Process | fruit dehiscence | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 342 aa Download sequence Send to blast |
MAENKVNLSI NGQSKVPPGF RFHPTEEELL HYYLRKKVNS QKIDLDVIRE VDLNKLEPWD 60 IQEECRIGST PQNDWYFFSH KDKKYPTGTR TNRATVAGFW KATGRDKIIY SCVRRIGLRK 120 TLVFYKGRAP HGQKSDWIMH EYRLDDTPAS NFHYQGSSDV ASEDPMSYNE DGWVVCRVFR 180 KKNYQKIDDC PKITISSSPD DTEQERTTFH NTQNDTGLDH ILLYMDRTSS SNFCNPESQT 240 LPTINIQHQH DLLFMQLPRL EETPPKSENP VQEKITGKPV CSNWGSLDRL VAWQLNNGHH 300 RMADTCDCPS FDEEEENGDN IDLWSSFTAS PSSLDPLVHL SV |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 3ulx_A | 1e-47 | 16 | 183 | 15 | 170 | Stress-induced transcription factor NAC1 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcriptional activator of genes involved in biosynthesis of secondary walls. Together with NST1, required for the secondary cell wall thickening and lignification of sclerenchymatous fibers and secondary xylem vessels (tracheary elements). Seems to repress the secondary cell wall thickening of xylary fibers. May also regulate the secondary cell wall lignification of other tissues. Binds to and activates the promoter of MYB46. {ECO:0000269|PubMed:17114348, ECO:0000269|PubMed:17237351, ECO:0000269|PubMed:17333250, ECO:0000269|PubMed:17565617, ECO:0000269|PubMed:17890373}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00016 | PBM | Transfer from AT1G32770 | Download |
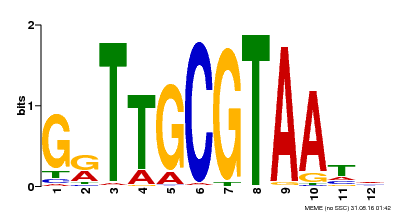
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | KFK45026.1 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | EF101892 | 0.0 | EF101892.1 Arabidopsis thaliana SND1 mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | NP_174554.1 | 0.0 | NAC domain containing protein 12 | ||||
| Swissprot | Q9LPI7 | 0.0 | NAC12_ARATH; NAC domain-containing protein 12 | ||||
| TrEMBL | A0A087HSC4 | 0.0 | A0A087HSC4_ARAAL; Uncharacterized protein | ||||
| STRING | A0A087HSC4 | 0.0 | (Arabis alpina) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM778 | 28 | 124 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G32770.1 | 0.0 | NAC domain containing protein 12 | ||||



