 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | KFK45057.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Arabideae; Arabis
|
||||||||
| Family | Trihelix | ||||||||
| Protein Properties | Length: 678aa MW: 75232 Da PI: 6.0947 | ||||||||
| Description | Trihelix family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | trihelix | 97.1 | 1.5e-30 | 55 | 139 | 1 | 87 |
trihelix 1 rWtkqevlaLiearremeerlrrgklkkplWeevskkmrergferspkqCkekwenlnkrykkikegekkrtsessstcpyfdqlea 87
rW+++e+laL+++r++m++++r++ lk+plWe+vs+k+ e g++rs+k+Ckek+en++k+yk++ke++++r++++ +++f+qlea
KFK45057.1 55 RWPREETLALLRIRSDMDTTFRDATLKAPLWEHVSRKLLELGYKRSAKKCKEKFENVQKYYKRTKETRGGRHDGK--AYKFFSQLEA 139
8*********************************************************************86555..5******985 PP
| |||||||
| 2 | trihelix | 104.2 | 9.3e-33 | 440 | 524 | 1 | 86 |
trihelix 1 rWtkqevlaLiearremeerlrrgklkkplWeevskkmrergferspkqCkekwenlnkrykkikegekkrtsessstcpyfdqle 86
rW+k e+laLi++r+ me r++++ k+ lWee+s m++ g++r++k+Ckekwen+nk+ykk+ke++kkr +++ +tcpyf++l+
KFK45057.1 440 RWPKAEILALINLRSGMEPRYQDNVPKGLLWEEISTSMKRMGYNRNAKRCKEKWENINKYYKKVKESNKKR-PQDAKTCPYFHRLD 524
8*********************************************************************8.99999*******97 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS50090 | 6.946 | 48 | 112 | IPR017877 | Myb-like domain |
| SMART | SM00717 | 0.0027 | 52 | 114 | IPR001005 | SANT/Myb domain |
| Pfam | PF13837 | 1.5E-20 | 54 | 140 | No hit | No description |
| CDD | cd12203 | 6.25E-26 | 54 | 119 | No hit | No description |
| SMART | SM00717 | 0.011 | 437 | 499 | IPR001005 | SANT/Myb domain |
| Pfam | PF13837 | 5.9E-22 | 439 | 524 | No hit | No description |
| PROSITE profile | PS50090 | 6.667 | 439 | 497 | IPR017877 | Myb-like domain |
| CDD | cd12203 | 1.30E-29 | 440 | 504 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0008361 | Biological Process | regulation of cell size | ||||
| GO:0010090 | Biological Process | trichome morphogenesis | ||||
| GO:0030308 | Biological Process | negative regulation of cell growth | ||||
| GO:0032876 | Biological Process | negative regulation of DNA endoreduplication | ||||
| GO:0042631 | Biological Process | cellular response to water deprivation | ||||
| GO:0045892 | Biological Process | negative regulation of transcription, DNA-templated | ||||
| GO:2000037 | Biological Process | regulation of stomatal complex patterning | ||||
| GO:2000038 | Biological Process | regulation of stomatal complex development | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 678 aa Download sequence Send to blast |
MEQAGGGGNE VVEEASPISS RPPANLEELM RFSAADEGGV GGGGGGSSSS SSGNRWPREE 60 TLALLRIRSD MDTTFRDATL KAPLWEHVSR KLLELGYKRS AKKCKEKFEN VQKYYKRTKE 120 TRGGRHDGKA YKFFSQLEAL NTSPPPPLLP PPPPSSSSLD ATPLSVANPP PSSSHHQFSV 180 FPQPQPPQQQ HITHTVSFTT PIPPLPPPLV APTFPGVAFS SHSSSTASGM GSDDEDDEDD 240 MDLDQAAGPS SRKRKRGNRG LGGGKMMELF EGLVRQVMQK QAAMQRSFLE ALEKREQERL 300 DREEAWKRQE MSRLAREHEV MSQERAASAS RDAAIISLIQ KITGHTVQLP PSLSSQPQPP 360 QPPPQTHQPP PAAKRVSTQA VVEQQHSTAQ SQSQPIMAIP QQQILPSPPP STLPPQQQDQ 420 KPQQEMIMSS EQSLSPSSSR WPKAEILALI NLRSGMEPRY QDNVPKGLLW EEISTSMKRM 480 GYNRNAKRCK EKWENINKYY KKVKESNKKR PQDAKTCPYF HRLDLLYRNK VLGSGGGGGV 540 ASSSGLQLQD QITSTVPQQC PVTAMKPPQE GVFNIHQPHH GSGSSEEEEP IEKSPQGTEK 600 PEDLVMRELI QQESMIGEYE KIEESHNYNN MEEDEEELDE EELDEDEKSA AYEIAFQSPT 660 NRGGNNGHTE PPFLTMVQ |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 97 | 105 | KRSAKKCKE |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription repressor that binds specific DNA sequence such as GT3 box 5'-GGTAAA-3' in the SDD1 promoter. Negative regulator of water use efficiency (WUE) via the promotion of stomatal density and distribution by the transcription repression of SDD1. Regulates the expression of several cell cycle genes and endoreduplication, especially in trichomes where it prevents ploidy-dependent plant cell growth. {ECO:0000269|PubMed:19717615, ECO:0000269|PubMed:21169508}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00177 | DAP | Transfer from AT1G33240 | Download |
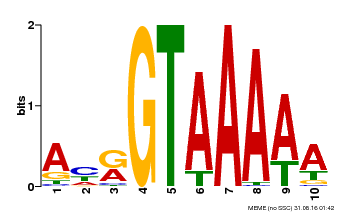
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | KFK45057.1 |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Down-regulated by water stress. {ECO:0000269|PubMed:21169508}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AC021045 | 1e-159 | AC021045.2 Arabidopsis thaliana chromosome I BAC T9L6 genomic sequence, complete sequence. | |||
| GenBank | AC027035 | 1e-159 | AC027035.5 Arabidopsis thaliana chromosome 1 BAC T16O9 genomic sequence, complete sequence. | |||
| GenBank | AJ003215 | 1e-159 | AJ003215.1 Arabidopsis thaliana GTL1 gene. | |||
| GenBank | CP002684 | 1e-159 | CP002684.1 Arabidopsis thaliana chromosome 1 sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_013742646.1 | 0.0 | trihelix transcription factor GTL1-like isoform X1 | ||||
| Swissprot | Q9C882 | 0.0 | GTL1_ARATH; Trihelix transcription factor GTL1 | ||||
| TrEMBL | A0A087HSF5 | 0.0 | A0A087HSF5_ARAAL; Uncharacterized protein | ||||
| STRING | A0A087HSF5 | 0.0 | (Arabis alpina) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM6226 | 27 | 46 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G33240.1 | 0.0 | GT-2-like 1 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



