 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | KHM99487.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Fabales; Fabaceae; Papilionoideae; Phaseoleae; Glycine; Soja
|
||||||||
| Family | WRKY | ||||||||
| Protein Properties | Length: 546aa MW: 59323.6 Da PI: 6.6532 | ||||||||
| Description | WRKY family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | WRKY | 101.1 | 6.4e-32 | 192 | 249 | 2 | 60 |
--SS-EEEEEEE--TT-SS-EEEEEE-STT---EEEEEE-SSSTTEEEEEEES--SS-- CS
WRKY 2 dDgynWrKYGqKevkgsefprsYYrCtsagCpvkkkversaedpkvveitYegeHnhek 60
dDgynWrKYGqK+vkgsefprsYY+Ct+++C+vkk ers+ d++++ei+Y+g+H+h+k
KHM99487.1 192 DDGYNWRKYGQKHVKGSEFPRSYYKCTHPNCEVKKLFERSH-DGQITEIIYKGTHDHPK 249
8****************************************.***************85 PP
| |||||||
| 2 | WRKY | 105.8 | 2.2e-33 | 366 | 424 | 1 | 59 |
---SS-EEEEEEE--TT-SS-EEEEEE-STT---EEEEEE-SSSTTEEEEEEES--SS- CS
WRKY 1 ldDgynWrKYGqKevkgsefprsYYrCtsagCpvkkkversaedpkvveitYegeHnhe 59
ldDgy+WrKYGqK+v+g+++prsYY+Ct +gCpv+k+ver+++dpk+v++tYeg+Hnh+
KHM99487.1 366 LDDGYRWRKYGQKVVRGNPNPRSYYKCTNTGCPVRKHVERASHDPKAVITTYEGKHNHD 424
59********************************************************7 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:2.20.25.80 | 1.1E-27 | 185 | 250 | IPR003657 | WRKY domain |
| PROSITE profile | PS50811 | 23.835 | 186 | 250 | IPR003657 | WRKY domain |
| SuperFamily | SSF118290 | 3.27E-25 | 188 | 250 | IPR003657 | WRKY domain |
| SMART | SM00774 | 4.1E-34 | 191 | 249 | IPR003657 | WRKY domain |
| Pfam | PF03106 | 1.6E-24 | 192 | 248 | IPR003657 | WRKY domain |
| Gene3D | G3DSA:2.20.25.80 | 1.4E-37 | 351 | 426 | IPR003657 | WRKY domain |
| SuperFamily | SSF118290 | 2.09E-29 | 358 | 426 | IPR003657 | WRKY domain |
| PROSITE profile | PS50811 | 38.664 | 361 | 426 | IPR003657 | WRKY domain |
| SMART | SM00774 | 3.5E-39 | 366 | 425 | IPR003657 | WRKY domain |
| Pfam | PF03106 | 4.1E-26 | 367 | 424 | IPR003657 | WRKY domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009611 | Biological Process | response to wounding | ||||
| GO:0009961 | Biological Process | response to 1-aminocyclopropane-1-carboxylic acid | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 546 aa Download sequence Send to blast |
MSPAKLPISR SPCVTIPPGL SPTSFLESPV LLSNMKVEPS PTTGSLSLLH QTAYGSMTSA 60 ASATFPVTTV CFNSNTVDER KPSFFEFKPH SGSNMVPADF DNHASEKSTQ IDSQGKAQAF 120 DSSALVKNES ASPSNELSLS SPVQMDCSGA SARVEGDLDE LNPRSNITTG LQASQVDNRG 180 SGLTVAAERV SDDGYNWRKY GQKHVKGSEF PRSYYKCTHP NCEVKKLFER SHDGQITEII 240 YKGTHDHPKP QPNRRYSAGT IMSVQEDRSD KASLTSRDDK GSNMCGQGSH LAEPDGKPEL 300 LPVATNDGDL DGLGVLSNRN NDEVDDDDPF SKRRKMDVGI ADITPVVKPI REPRVVVQTL 360 SEVDILDDGY RWRKYGQKVV RGNPNPRSYY KCTNTGCPVR KHVERASHDP KAVITTYEGK 420 HNHDVPTARN SCHDMAGPAS ASGQTRVRPE ESDTISLDLG MGISPAAENT SNSQGRMMLS 480 EFGDSQIHTS NSNFKFVHTT TAPGYFGVLN NNSNPYGSKE NPSDGPSLNH SAYPCPQNIG 540 RILMGP |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1wj2_A | 1e-36 | 181 | 426 | 6 | 77 | Probable WRKY transcription factor 4 |
| 2lex_A | 1e-36 | 181 | 426 | 6 | 77 | Probable WRKY transcription factor 4 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription factor. Interacts specifically with the W box (5'-(T)TGAC[CT]-3'), a frequently occurring elicitor-responsive cis-acting element. {ECO:0000250|UniProtKB:Q9SI37}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00455 | DAP | Transfer from AT4G26640 | Download |
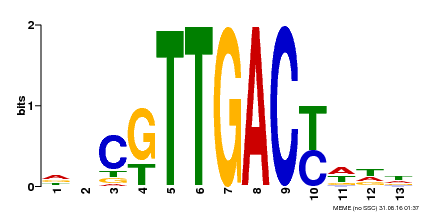
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | KHM99487.1 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | EU019574 | 0.0 | EU019574.1 Glycine max WRKY35 (WRKY35) mRNA, partial cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_003530784.1 | 0.0 | probable WRKY transcription factor 20 isoform X1 | ||||
| Refseq | XP_028246070.1 | 0.0 | probable WRKY transcription factor 20 isoform X1 | ||||
| Swissprot | Q93WV0 | 0.0 | WRK20_ARATH; Probable WRKY transcription factor 20 | ||||
| TrEMBL | A0A445JNA3 | 0.0 | A0A445JNA3_GLYSO; Putative WRKY transcription factor 20 isoform A | ||||
| TrEMBL | I1KYG7 | 0.0 | I1KYG7_SOYBN; Uncharacterized protein | ||||
| STRING | GLYMA08G43770.1 | 0.0 | (Glycine max) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF3946 | 34 | 63 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G26640.2 | 0.0 | WRKY family protein | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



