 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | KHN00315.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Fabales; Fabaceae; Papilionoideae; Phaseoleae; Glycine; Soja
|
||||||||
| Family | SBP | ||||||||
| Protein Properties | Length: 957aa MW: 106537 Da PI: 6.1791 | ||||||||
| Description | SBP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | SBP | 128.8 | 2.1e-40 | 99 | 176 | 1 | 78 |
--SSTT-----TT--HHHHHTT--HHHHT-S-EEETTEEEEE-TTTSSEEETTT--SS--S-STTTT-------S--- CS
SBP 1 lCqvegCeadlseakeyhrrhkvCevhskapvvlvsgleqrfCqqCsrfhelsefDeekrsCrrrLakhnerrrkkqa 78
+Cqve+C+adls+ak+yhrrhkvCe+hska+ +lv + +qrfCqqCsrfh l+efDe+krsCrrrLa+hn+rrrk+++
KHN00315.1 99 VCQVEDCSADLSKAKDYHRRHKVCEMHSKASRALVGNAMQRFCQQCSRFHLLQEFDEGKRSCRRRLAGHNKRRRKTNH 176
6**************************************************************************976 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:4.10.1100.10 | 3.1E-33 | 93 | 161 | IPR004333 | Transcription factor, SBP-box |
| PROSITE profile | PS51141 | 32.18 | 97 | 174 | IPR004333 | Transcription factor, SBP-box |
| SuperFamily | SSF103612 | 9.94E-38 | 98 | 178 | IPR004333 | Transcription factor, SBP-box |
| Pfam | PF03110 | 3.5E-29 | 100 | 173 | IPR004333 | Transcription factor, SBP-box |
| Gene3D | G3DSA:1.25.40.20 | 2.1E-6 | 706 | 842 | IPR020683 | Ankyrin repeat-containing domain |
| SuperFamily | SSF48403 | 1.38E-7 | 728 | 841 | IPR020683 | Ankyrin repeat-containing domain |
| CDD | cd00204 | 1.69E-6 | 731 | 842 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0016021 | Cellular Component | integral component of membrane | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 957 aa Download sequence Send to blast |
MVLEWDSNFS LGILVAGGPS NSSSTSEEVD PRDPKGNKEG DKKRRVIVLE DDGLNEEGGT 60 LSLKLGGHAS AVVDREVGSW DGTNGKKSRV SGSTSNRAVC QVEDCSADLS KAKDYHRRHK 120 VCEMHSKASR ALVGNAMQRF CQQCSRFHLL QEFDEGKRSC RRRLAGHNKR RRKTNHEAVP 180 NGSSLNDDQT SSYLLISLLK ILSNMHSDRS DQTTDQDLLT HILRSLASQN GEQGSKNIAN 240 LLREPENLLR EDGSSRKSEM MSTLFSNGSQ GSPTDTRQHE TVSIAKMQQQ VMHAHDARAA 300 DQQITSSIKP SMSNSPPAYS EARDSTAGQI KMNNFDLNDI YIDSDDGMED LERLPVSTNL 360 VTSSLDYPWA QQDSHQSSPP QTSGNSDSAS AQSPSSFSGE AQSRTDRIVF KLFGKEPNDF 420 PLVLRAQILD WLSHSPTDME SYIRPGCIVL TIYLRQAEAL WEELCYDLTS SLNRLLDVSD 480 DTFWRNGWVH IRVQHQMAFI FNGQVVIDTS LPFRSNNYSK ILTVSPIAAP ASKRAQFSVK 540 GVNLIRPATR LMCALEGKYL VCEDAHMSMD QSSKEPDELQ CVQFSCSVPV MNGRGFIEIE 600 DQGLSSSFFP FIVVEEDVCS EICTLEPLLE LSETDPDIEG TGKIKAKNQA MDFIHEMGWL 660 LHRSQLKLRM VQLNSSEDLF PLKRFKWLIE FSMDHDWCAA VRKLLNLLLD GTVNTGDHPS 720 LYLALSEMGL LHKAVRRNSK QLVECLLRYV PENISDKLGP EDKALVDGEN QTFLFRPDVV 780 GPAGLTPLHI AAGKDGSEDV LDALTNDPCM VGIEAWKNAR DSTGSTPEDY ARLRGHYAYI 840 HLVQKKINKR QGAAHVVVEI PSNTTESNTN EKQNELSTTF EIGKAEVIRG QGHCKLCDKR 900 ISCRTAVGRS LVYRPAMLSM VAIAAVCVCV ALLFKSSPEV ICMFRPFRWE NLDFGTS |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1ul4_A | 6e-31 | 91 | 173 | 3 | 84 | squamosa promoter binding protein-like 4 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
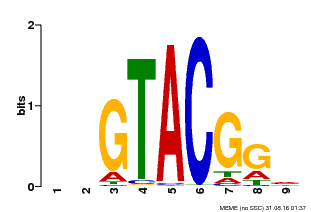
| UniProt | Trans-acting factor that binds specifically to the consensus nucleotide sequence 5'-TNCGTACAA-3'. {ECO:0000269|PubMed:16554053}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00633 | PBM | Transfer from PK06791.1 | Download |
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | KHN00315.1 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | NP_001352074.1 | 0.0 | squamosa promoter-binding-like protein 1 | ||||
| Refseq | XP_028184385.1 | 0.0 | squamosa promoter-binding-like protein 1 | ||||
| Swissprot | Q9S7P5 | 0.0 | SPL12_ARATH; Squamosa promoter-binding-like protein 12 | ||||
| TrEMBL | A0A445IG44 | 0.0 | A0A445IG44_GLYSO; Squamosa promoter-binding-like protein 1 isoform A | ||||
| STRING | GLYMA10G01211.1 | 0.0 | (Glycine max) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF1539 | 34 | 95 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G60030.1 | 0.0 | squamosa promoter-binding protein-like 12 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



