 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | KHN02156.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Fabales; Fabaceae; Papilionoideae; Phaseoleae; Glycine; Soja
|
||||||||
| Family | MYB | ||||||||
| Protein Properties | Length: 309aa MW: 35146.7 Da PI: 9.3995 | ||||||||
| Description | MYB family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 23.6 | 1.2e-07 | 28 | 73 | 3 | 46 |
SS-HHHHHHHHHHHHHTTTT...-HHHHHHHHTTTS-HHHHHHHHHH CS
Myb_DNA-binding 3 rWTteEdellvdavkqlGgg...tWktIartmgkgRtlkqcksrwqk 46
rWT++E +l+ +a + + ++ +W ++a+ ++ g+t +++ ++
KHN02156.1 28 RWTPQENKLFENALAVFDKDtpdRWLKVAALIP-GKTVGDVIKQYRE 73
9********************************.******9999986 PP
| |||||||
| 2 | Myb_DNA-binding | 46.6 | 8.1e-15 | 136 | 180 | 3 | 47 |
SS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHH CS
Myb_DNA-binding 3 rWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqky 47
+WT+eE+ +++ + k++G+g+W+ I+r + ++Rt+ q+ s+ qky
KHN02156.1 136 PWTKEEHRQFLMGLKKYGKGDWRNISRNFVTTRTPTQVASHAQKY 180
8*******************************************9 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51293 | 9.629 | 24 | 79 | IPR017884 | SANT domain |
| SMART | SM00717 | 1.3E-7 | 25 | 77 | IPR001005 | SANT/Myb domain |
| SuperFamily | SSF46689 | 4.38E-11 | 27 | 83 | IPR009057 | Homeodomain-like |
| CDD | cd00167 | 2.30E-6 | 28 | 74 | No hit | No description |
| Pfam | PF00249 | 6.2E-6 | 28 | 73 | IPR001005 | SANT/Myb domain |
| PROSITE profile | PS51294 | 20.236 | 129 | 185 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 2.23E-17 | 130 | 184 | IPR009057 | Homeodomain-like |
| TIGRFAMs | TIGR01557 | 4.5E-17 | 132 | 184 | IPR006447 | Myb domain, plants |
| SMART | SM00717 | 2.6E-13 | 133 | 183 | IPR001005 | SANT/Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 4.9E-13 | 133 | 180 | IPR009057 | Homeodomain-like |
| Pfam | PF00249 | 3.7E-12 | 136 | 180 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 4.70E-11 | 136 | 181 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 309 aa Download sequence Send to blast |
MNRGIEVLSP ASYLRSSNWL FQESLGTRWT PQENKLFENA LAVFDKDTPD RWLKVAALIP 60 GKTVGDVIKQ YRELEEDVSV IESGFIPLPG YTAADSFTLE WVNNQGFDGL RQFYGVTGKR 120 GASNRPSEQE RKKGVPWTKE EHRQFLMGLK KYGKGDWRNI SRNFVTTRTP TQVASHAQKY 180 FIRQLSGGKD KKRSSIHDIT MVNLPEAKSP SSESNNRPYS PDHSVKDVNP PSQNQRLSTT 240 MVIKQEHDWK LPCETVPMLF NSTNGNMFMT PLCGISSYGS KPLQEQYFLS GCQFGPYDTI 300 MQMPSMQNQ |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 2cjj_A | 3e-16 | 26 | 91 | 8 | 73 | RADIALIS |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Involved in the dorsovental asymmetry of flowers. Promotes ventral identity. {ECO:0000269|PubMed:11937495, ECO:0000269|PubMed:9118809}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00296 | DAP | Transfer from AT2G38090 | Download |
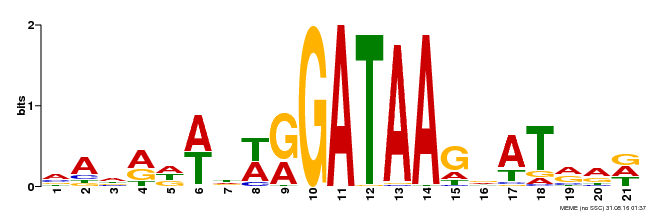
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | KHN02156.1 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | KT031184 | 0.0 | KT031184.1 Glycine max clone HN_CCL_88 MYB/HD-like transcription factor (Glyma03g14440.1) mRNA, partial cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | NP_001240137.1 | 0.0 | MYB transcription factor MYB51 | ||||
| Refseq | XP_006576272.1 | 0.0 | MYB transcription factor MYB51 isoform X1 | ||||
| Refseq | XP_014628871.1 | 0.0 | MYB transcription factor MYB51 isoform X1 | ||||
| Refseq | XP_028224761.1 | 0.0 | transcription factor DIVARICATA-like | ||||
| Refseq | XP_028224762.1 | 0.0 | transcription factor DIVARICATA-like | ||||
| Swissprot | Q8S9H7 | 1e-114 | DIV_ANTMA; Transcription factor DIVARICATA | ||||
| TrEMBL | A0A0B2NYD1 | 0.0 | A0A0B2NYD1_GLYSO; Transcription factor MYB1R1 | ||||
| STRING | GLYMA03G14440.1 | 0.0 | (Glycine max) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF466 | 34 | 162 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G38090.1 | 1e-123 | MYB family protein | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



