 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | KHN04852.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Fabales; Fabaceae; Papilionoideae; Phaseoleae; Glycine; Soja
|
||||||||
| Family | G2-like | ||||||||
| Protein Properties | Length: 375aa MW: 42117 Da PI: 9.5457 | ||||||||
| Description | G2-like family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | G2-like | 102.2 | 3.4e-32 | 194 | 247 | 2 | 55 |
G2-like 2 prlrWtpeLHerFveaveqLGGsekAtPktilelmkvkgLtlehvkSHLQkYRl 55
pr+rWt++LH+rFv+ave LGG+e+AtPk++lelm+vk+Ltl+hvkSHLQ+YR+
KHN04852.1 194 PRMRWTSTLHARFVHAVELLGGHERATPKSVLELMDVKDLTLAHVKSHLQMYRT 247
9****************************************************7 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF46689 | 1.97E-15 | 191 | 247 | IPR009057 | Homeodomain-like |
| Gene3D | G3DSA:1.10.10.60 | 1.4E-28 | 192 | 247 | IPR009057 | Homeodomain-like |
| TIGRFAMs | TIGR01557 | 8.8E-24 | 194 | 247 | IPR006447 | Myb domain, plants |
| Pfam | PF00249 | 6.4E-7 | 195 | 246 | IPR001005 | SANT/Myb domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009944 | Biological Process | polarity specification of adaxial/abaxial axis | ||||
| GO:0048481 | Biological Process | plant ovule development | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 375 aa Download sequence Send to blast |
MELFPAQPDL SLQISPPNAN PTSSWRRSTE EDMDLGFWKR ALDSRNSIQS TMAKQDSCFD 60 LSLSNPKASD NNNSNLIHHH HFQTSNATTI INPFQLPFQQ NHFFHQQQQP LFQPQHQSLS 120 QDLGFLRPIR GIPVYQNPPP IPFTQHHHLP LEASTTTPSI ISNTNTGSTP FHSQALMRSR 180 FLSRFPAKRS MRAPRMRWTS TLHARFVHAV ELLGGHERAT PKSVLELMDV KDLTLAHVKS 240 HLQMYRTVKT TDRAAASSGQ SDVYDNGSSG DTSDDLMFDI KSSRRSDLSV KQGRSSVNQD 300 KKYHGLWGNS SREAWLHGKT KADSVGNVPS YLEKEMDPKC LSYERISDGS SSSNLSGSSP 360 KKPNLDLEFS LGQPL |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 6j4k_A | 5e-17 | 195 | 249 | 4 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4k_B | 5e-17 | 195 | 249 | 4 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_A | 5e-17 | 195 | 249 | 4 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_C | 5e-17 | 195 | 249 | 4 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_D | 5e-17 | 195 | 249 | 4 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_F | 5e-17 | 195 | 249 | 4 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_H | 5e-17 | 195 | 249 | 4 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_J | 5e-17 | 195 | 249 | 4 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcription factor that regulates lateral organ polarity. Promotes abaxial cell fate during lateral organd formation. Functions with KAN1 in the specification of polarity of the ovule outer integument. {ECO:0000269|PubMed:11525739, ECO:0000269|PubMed:15286295, ECO:0000269|PubMed:16623911, ECO:0000269|PubMed:17307928}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00174 | DAP | Transfer from AT1G32240 | Download |
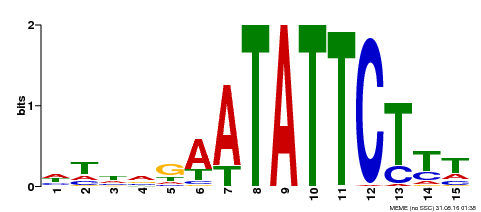
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | KHN04852.1 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_028227361.1 | 0.0 | probable transcription factor KAN2 | ||||
| Swissprot | Q9C616 | 3e-96 | KAN2_ARATH; Probable transcription factor KAN2 | ||||
| TrEMBL | A0A0B2PB80 | 0.0 | A0A0B2PB80_GLYSO; Putative transcription factor KAN2 | ||||
| STRING | XP_007158256.1 | 0.0 | (Phaseolus vulgaris) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF5968 | 34 | 52 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G32240.1 | 1e-76 | G2-like family protein | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



