 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | KHN04943.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Fabales; Fabaceae; Papilionoideae; Phaseoleae; Glycine; Soja
|
||||||||
| Family | MYB | ||||||||
| Protein Properties | Length: 963aa MW: 108507 Da PI: 5.0153 | ||||||||
| Description | MYB family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 43.1 | 9.6e-14 | 8 | 53 | 2 | 48 |
SSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHHT CS
Myb_DNA-binding 2 grWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqkyl 48
g W Ede+l+ av ++G+++W++I++ + ++++kqck rw+ +l
KHN04943.1 8 GVWKNTEDEILKAAVMKYGKNQWARISSLLV-RKSAKQCKARWYEWL 53
78*****************************.************986 PP
| |||||||
| 2 | Myb_DNA-binding | 32.8 | 1.6e-10 | 60 | 103 | 2 | 48 |
SSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHHT CS
Myb_DNA-binding 2 grWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqkyl 48
WT eEde+l+++ k++++ W+tIa +g Rt+ qc +r+ k+l
KHN04943.1 60 TEWTREEDEKLLHLAKLMPTQ-WRTIAPIVG--RTPSQCLERYEKLL 103
68*****************99.********8..**********9986 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51294 | 22.008 | 2 | 57 | IPR017930 | Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 5.4E-18 | 5 | 56 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 7.0E-15 | 6 | 55 | IPR001005 | SANT/Myb domain |
| Pfam | PF13921 | 4.1E-13 | 10 | 70 | No hit | No description |
| CDD | cd00167 | 7.30E-12 | 10 | 53 | No hit | No description |
| SuperFamily | SSF46689 | 8.27E-21 | 33 | 106 | IPR009057 | Homeodomain-like |
| CDD | cd11659 | 1.48E-30 | 55 | 106 | No hit | No description |
| Gene3D | G3DSA:1.10.10.60 | 3.1E-14 | 57 | 104 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS51294 | 16.443 | 58 | 107 | IPR017930 | Myb domain |
| SMART | SM00717 | 1.7E-12 | 58 | 105 | IPR001005 | SANT/Myb domain |
| Pfam | PF11831 | 1.3E-61 | 406 | 654 | IPR021786 | Pre-mRNA splicing factor component Cdc5p/Cef1 |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009870 | Biological Process | defense response signaling pathway, resistance gene-dependent | ||||
| GO:0010204 | Biological Process | defense response signaling pathway, resistance gene-independent | ||||
| GO:0042742 | Biological Process | defense response to bacterium | ||||
| GO:0050832 | Biological Process | defense response to fungus | ||||
| GO:0051301 | Biological Process | cell division | ||||
| GO:0009507 | Cellular Component | chloroplast | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 963 aa Download sequence Send to blast |
MRIMIKGGVW KNTEDEILKA AVMKYGKNQW ARISSLLVRK SAKQCKARWY EWLDPSIKKT 60 EWTREEDEKL LHLAKLMPTQ WRTIAPIVGR TPSQCLERYE KLLDAACVKD ENYEPGDDPR 120 KLRPGEIDPN PESKPARPDP VDMDEDEKEM LSEARARLAN TKGKKAKRKA REKQLEEARR 180 LASLQKKREL KAAGIDIRQR KRKRKGIDYN AEIPFEKRPP PGFFDVTDED RPVEQPQFPT 240 TIEELEGKRR VDVEAQLRKQ DIAKNKIAQR QDAPSAILHA NKLNDPETVR KRSKLMLPPP 300 QISDQELDEI AKLGYASDLA GSQELAEGSG ATRALLADYA QTPGQGMTPL RTPQRTPAGK 360 GDAIMMEAEN LARLRESQTP LLGGENPELH PSDFNGVTPK KKEIQTPNPM LTPSATPGGA 420 GLTPRIGMTP TRDGFSFSMT PKGTPLRDEL HINEDMNMHD STKLELQRQA DMRRSLRSGL 480 GSLPQPKNEY QIVMQPVPED AEEPEEKIEE DMSDRIAREK AEEEARQQAL LRKRSKVLQR 540 ELPRPPTASL ELIRNSLMRT DVDKSSFVPP TSIEQADEMI RRELLSLLEH DNAKYPLDEK 600 VIKEKKKGAK RAVNGSAVPV IEDFEEDEMK EADKLIKEEA LYLCAAMGHE DEPLDEFIEA 660 HRTCLNDLMY FPTRNAYGLS SVAGNMEKLT ALQNEFENVR SKLDDDKEKM VRLEKKVMVL 720 TQGYEMRVKK SLWPQIEATF KQMDVAATEL ECFKALQKQE QLAASHRINN LWAEVQKQKE 780 LEKTLQNRYG SLIEELEKMQ NVMDQCRLQA QQQEEIKANH ARESTETPET KADGIDVQGT 840 ANCEAVPHSV EHGRALAVES SADGTADQQV DIVHDQATSS VSHDMDVDSD KLANPTPAAE 900 NVDEKVEGTG TGTGSYTDDG ETMLEMGAAV EVSSPNHDVV VDAVNSHDNN SMEETNAVGE 960 ETY |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 5mqf_L | 0.0 | 2 | 801 | 3 | 800 | Cell division cycle 5-like protein |
| 5xjc_L | 0.0 | 2 | 801 | 3 | 800 | Cell division cycle 5-like protein |
| 5yzg_L | 0.0 | 2 | 801 | 3 | 800 | Cell division cycle 5-like protein |
| 5z56_L | 0.0 | 2 | 801 | 3 | 800 | Cell division cycle 5-like protein |
| 5z57_L | 0.0 | 2 | 801 | 3 | 800 | Cell division cycle 5-like protein |
| 5z58_L | 0.0 | 2 | 801 | 3 | 800 | Cell division cycle 5-like protein |
| 6ff4_L | 0.0 | 2 | 801 | 3 | 800 | Cell division cycle 5-like protein |
| 6ff7_L | 0.0 | 2 | 801 | 3 | 800 | Cell division cycle 5-like protein |
| 6icz_L | 0.0 | 2 | 801 | 3 | 800 | Cell division cycle 5-like protein |
| 6id0_L | 0.0 | 2 | 801 | 3 | 800 | Cell division cycle 5-like protein |
| 6id1_L | 0.0 | 2 | 801 | 3 | 800 | Cell division cycle 5-like protein |
| 6qdv_O | 0.0 | 2 | 801 | 3 | 800 | Cell division cycle 5-like protein |
| Search in ModeBase | ||||||
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 185 | 204 | KKRELKAAGIDIRQRKRKRK |
| 2 | 199 | 204 | RKRKRK |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Component of the MAC complex that probably regulates defense responses through transcriptional control and thereby is essential for plant innate immunity. Possesses a sequence specific DNA sequence 'CTCAGCG' binding activity. Involved in mRNA splicing and cell cycle control. May also play a role in the response to DNA damage. {ECO:0000250|UniProtKB:Q99459, ECO:0000269|PubMed:17298883, ECO:0000269|PubMed:17575050, ECO:0000269|PubMed:8917598}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00028 | SELEX | Transfer from AT1G09770 | Download |
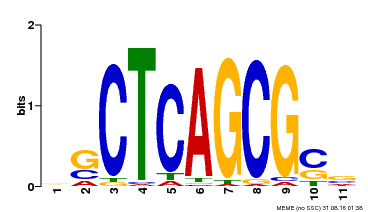
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | KHN04943.1 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AP015044 | 0.0 | AP015044.1 Vigna angularis var. angularis DNA, chromosome 11, almost complete sequence, cultivar: Shumari. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_028221963.1 | 0.0 | cell division cycle 5-like protein isoform X1 | ||||
| Swissprot | P92948 | 0.0 | CDC5L_ARATH; Cell division cycle 5-like protein | ||||
| TrEMBL | A0A445F8W3 | 0.0 | A0A445F8W3_GLYSO; Cell division cycle 5-like protein | ||||
| STRING | GLYMA20G36600.2 | 0.0 | (Glycine max) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF4072 | 33 | 54 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G09770.1 | 0.0 | cell division cycle 5 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



