 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | KHN13627.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Fabales; Fabaceae; Papilionoideae; Phaseoleae; Glycine; Soja
|
||||||||
| Family | HD-ZIP | ||||||||
| Protein Properties | Length: 268aa MW: 30488.3 Da PI: 7.6486 | ||||||||
| Description | HD-ZIP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Homeobox | 58.7 | 9.9e-19 | 80 | 132 | 4 | 56 |
-SS--HHHHHHHHHHHHHSSS--HHHHHHHHHHCTS-HHHHHHHHHHHHHHHH CS
Homeobox 4 RttftkeqleeLeelFeknrypsaeereeLAkklgLterqVkvWFqNrRakek 56
+ +++ eq+++Le+ F + +++ e++ +LAk lgL+ rqV +WFqNrRa++k
KHN13627.1 80 KKRLNLEQVKALEKSFDQGNKLEPERKVQLAKALGLQPRQVAIWFQNRRARWK 132
4568889*********************************************9 PP
| |||||||
| 2 | HD-ZIP_I/II | 126 | 1.7e-40 | 78 | 170 | 1 | 93 |
HD-ZIP_I/II 1 ekkrrlskeqvklLEesFeeeekLeperKvelareLglqprqvavWFqnrRARtktkqlEkdyeaLkraydalkeenerLekeveeLreelke 93
ekk+rl+ eqvk+LE+sF++ +kLeperKv+la++Lglqprqva+WFqnrRAR+ktkqlEk+ye+Lk++++a+k++n+ L+ +++L++el++
KHN13627.1 78 EKKKRLNLEQVKALEKSFDQGNKLEPERKVQLAKALGLQPRQVAIWFQNRRARWKTKQLEKEYEVLKKQFEAVKADNDVLKVRNQKLQAELQA 170
69***************************************************************************************9986 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF46689 | 1.8E-19 | 70 | 136 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS50071 | 17.07 | 74 | 134 | IPR001356 | Homeobox domain |
| SMART | SM00389 | 4.0E-18 | 77 | 138 | IPR001356 | Homeobox domain |
| CDD | cd00086 | 2.07E-16 | 79 | 135 | No hit | No description |
| Pfam | PF00046 | 5.6E-16 | 80 | 132 | IPR001356 | Homeobox domain |
| Gene3D | G3DSA:1.10.10.60 | 3.1E-19 | 86 | 133 | IPR009057 | Homeodomain-like |
| PRINTS | PR00031 | 7.2E-6 | 105 | 114 | IPR000047 | Helix-turn-helix motif |
| PROSITE pattern | PS00027 | 0 | 109 | 132 | IPR017970 | Homeobox, conserved site |
| PRINTS | PR00031 | 7.2E-6 | 114 | 130 | IPR000047 | Helix-turn-helix motif |
| Pfam | PF02183 | 1.1E-11 | 134 | 173 | IPR003106 | Leucine zipper, homeobox-associated |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009733 | Biological Process | response to auxin | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 268 aa Download sequence Send to blast |
MAFLPSHGFT FNTTHEEDHC HFPPTSLDAF PSLPPQHFQG GAPFMLKRSM SLSGIENKCN 60 EVHGDDELSD DGVFQSGEKK KRLNLEQVKA LEKSFDQGNK LEPERKVQLA KALGLQPRQV 120 AIWFQNRRAR WKTKQLEKEY EVLKKQFEAV KADNDVLKVR NQKLQAELQA VKSRDCCEAG 180 IISLKKETEG SWSNGSDNNS DLNLDPSRAL GLNSPISSHN IKNLLPNSLK PNSITQLLQD 240 DGLCNMFHNI DAPQNFWPRP DQHHQRFH |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 126 | 134 | RRARWKTKQ |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcription factor. {ECO:0000250}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00322 | DAP | Transfer from AT3G01220 | Download |
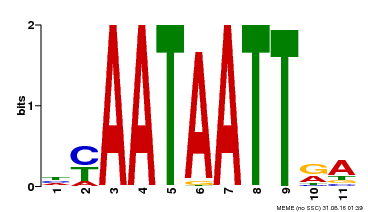
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | KHN13627.1 |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: By auxin. {ECO:0000269|PubMed:12644682}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AC235119 | 1e-161 | AC235119.1 Glycine max strain Williams 82 clone GM_WBa0029B22, complete sequence. | |||
| GenBank | AC235240 | 1e-161 | AC235240.1 Glycine max strain Williams 82 clone GM_WBb0029M05, complete sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_028213077.1 | 0.0 | homeobox-leucine zipper protein ATHB-20-like | ||||
| Swissprot | Q8LAT0 | 2e-81 | ATB20_ARATH; Homeobox-leucine zipper protein ATHB-20 | ||||
| TrEMBL | A0A445FXU0 | 0.0 | A0A445FXU0_GLYSO; Homeobox-leucine zipper protein ATHB-20 isoform A | ||||
| STRING | GLYMA18G49290.1 | 0.0 | (Glycine max) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF1271 | 33 | 106 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G01220.1 | 1e-75 | homeobox protein 20 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



