 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | KHN18811.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Fabales; Fabaceae; Papilionoideae; Phaseoleae; Glycine; Soja
|
||||||||
| Family | MYB_related | ||||||||
| Protein Properties | Length: 332aa MW: 36845.7 Da PI: 9.1996 | ||||||||
| Description | MYB_related family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 42.4 | 1.6e-13 | 59 | 103 | 1 | 47 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHH CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqky 47
r +WT++E++++++a +++ + Wk+I + +g +t q++s+ qky
KHN18811.1 59 RESWTEQEHDKFLEALQLFDRD-WKKIEAFVG-SKTVIQIRSHAQKY 103
789*****************77.*********.*************9 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF46689 | 1.05E-16 | 53 | 109 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS51294 | 15.408 | 54 | 108 | IPR017930 | Myb domain |
| TIGRFAMs | TIGR01557 | 1.5E-18 | 57 | 106 | IPR006447 | Myb domain, plants |
| Gene3D | G3DSA:1.10.10.60 | 1.1E-6 | 57 | 109 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 5.3E-11 | 58 | 106 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 2.8E-11 | 59 | 103 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 2.06E-8 | 61 | 104 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009651 | Biological Process | response to salt stress | ||||
| GO:0009723 | Biological Process | response to ethylene | ||||
| GO:0009733 | Biological Process | response to auxin | ||||
| GO:0009737 | Biological Process | response to abscisic acid | ||||
| GO:0009739 | Biological Process | response to gibberellin | ||||
| GO:0009751 | Biological Process | response to salicylic acid | ||||
| GO:0009753 | Biological Process | response to jasmonic acid | ||||
| GO:0046686 | Biological Process | response to cadmium ion | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 332 aa Download sequence Send to blast |
MVSVNPNPAQ GFYFFDPSNM TLPGVNNLPP PPPPAPAAPS AVEDPNKKIR KPYTITKSRE 60 SWTEQEHDKF LEALQLFDRD WKKIEAFVGS KTVIQIRSHA QKYFLKVQKN GTSEHVPPPR 120 PKRKAARPYP QKAPKTPTVS QVMGPLQSSS AFIEPAYIYS PDSSSVLGTP VTNMPLSSWN 180 YNTTPQPGNV PQVTRDDMGL TGAGQAAPLN CCYSSSNEST PPTWPRSKRI NQGDQDKPIK 240 VMPDFAQVYS FIGSVFDPNS TNHLQKLQQM DPINVETVLL LMTNLSVNLM SPEFEDHKRL 300 LSSYDTDSDK SKFVNICSKS FTNKSESAVL SA |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcription factor. {ECO:0000250}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00423 | DAP | Transfer from AT4G01280 | Download |
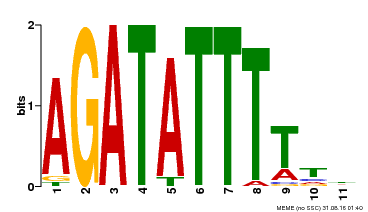
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | KHN18811.1 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_028206882.1 | 0.0 | protein REVEILLE 5-like isoform X1 | ||||
| Swissprot | C0SVG5 | 1e-110 | RVE5_ARATH; Protein REVEILLE 5 | ||||
| TrEMBL | A0A0B2QB51 | 0.0 | A0A0B2QB51_GLYSO; Protein REVEILLE 5 isoform A | ||||
| STRING | GLYMA16G03640.1 | 0.0 | (Glycine max) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF1119 | 33 | 96 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G01280.1 | 1e-95 | MYB_related family protein | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



