 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | KHN23589.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Fabales; Fabaceae; Papilionoideae; Phaseoleae; Glycine; Soja
|
||||||||
| Family | MYB_related | ||||||||
| Protein Properties | Length: 466aa MW: 51784.9 Da PI: 5.5919 | ||||||||
| Description | MYB_related family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 53.3 | 6.3e-17 | 46 | 90 | 1 | 47 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHH CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqky 47
r +WT+eE++++++a k++G g W+ I +++g ++t+ q++s+ qk+
KHN23589.1 46 REKWTEEEHQKFLEALKLYGRG-WRQIEEHIG-TKTAVQIRSHAQKF 90
789*******************.*********.************98 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF46689 | 3.81E-16 | 40 | 94 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS51294 | 21.201 | 41 | 95 | IPR017930 | Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 5.0E-10 | 43 | 99 | IPR009057 | Homeodomain-like |
| TIGRFAMs | TIGR01557 | 3.7E-16 | 44 | 93 | IPR006447 | Myb domain, plants |
| SMART | SM00717 | 4.5E-13 | 45 | 93 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 1.2E-14 | 46 | 89 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 2.57E-11 | 48 | 91 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0007623 | Biological Process | circadian rhythm | ||||
| GO:0009651 | Biological Process | response to salt stress | ||||
| GO:0009723 | Biological Process | response to ethylene | ||||
| GO:0046686 | Biological Process | response to cadmium ion | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 466 aa Download sequence Send to blast |
MEMQDQIEST RSTIFGSASN IHSNAEKQAE NVAPKVRKPY TITKQREKWT EEEHQKFLEA 60 LKLYGRGWRQ IEEHIGTKTA VQIRSHAQKF FSKVVRESEV SDEGSIQPIN IPPPRPKRKP 120 LHPYPRKSVN SFRGPTIPNE TEISPSTNLL VAEKDTPSPT SVLSTVGSEA FGSQFSEQTN 180 RCLSPNSCTT DIHSVSLSPV EKENDCMTSK ESEEEEKASP ASRPLSTVSN PKMCMKPEFS 240 SKEIEDATDM PQTTSIKLFG RTVSMVGNQK SLKIDDDGKP ITVKSDEVDD VENEKLGQSG 300 ESKQVDTQLS LGVVSGNWPI TPDADGANVT SIEPPKENLC FSESAPDAFF PQWSLSQGLP 360 PFYLRPCNQV LNPLPLRPSS LKVRTREEES CCTGSNTESV CDMENQGKNS DAVDSKFQKY 420 HEEGAAPQKK PARGFVPYKR CLAERDGHSF IVAMEEREGQ RARVCS |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00146 | DAP | Transfer from AT1G18330 | Download |
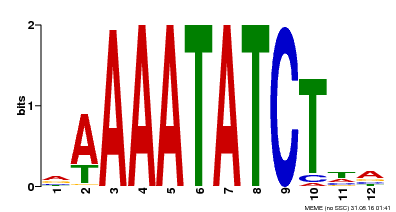
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | KHN23589.1 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_003534089.1 | 0.0 | protein REVEILLE 7-like | ||||
| Refseq | XP_028181748.1 | 0.0 | protein REVEILLE 7-like | ||||
| TrEMBL | A0A0B2QU28 | 0.0 | A0A0B2QU28_GLYSO; Protein LHY | ||||
| TrEMBL | I1L3Y3 | 0.0 | I1L3Y3_SOYBN; Uncharacterized protein | ||||
| STRING | GLYMA09G29800.3 | 0.0 | (Glycine max) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF7313 | 32 | 44 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G18330.1 | 4e-53 | MYB_related family protein | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



