 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | KHN27648.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Fabales; Fabaceae; Papilionoideae; Phaseoleae; Glycine; Soja
|
||||||||
| Family | G2-like | ||||||||
| Protein Properties | Length: 420aa MW: 47416.1 Da PI: 8.0102 | ||||||||
| Description | G2-like family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | G2-like | 106.1 | 2e-33 | 46 | 100 | 1 | 55 |
G2-like 1 kprlrWtpeLHerFveaveqLGGsekAtPktilelmkvkgLtlehvkSHLQkYRl 55
kprl+Wtp+LH+rF+eav+qLGG++kAtPkt+++lm+++gLtl+h+kSHLQkYRl
KHN27648.1 46 KPRLKWTPDLHARFIEAVNQLGGADKATPKTVMKLMGIPGLTLYHLKSHLQKYRL 100
79****************************************************8 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51294 | 10.489 | 43 | 103 | IPR017930 | Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 4.4E-31 | 44 | 101 | IPR009057 | Homeodomain-like |
| SuperFamily | SSF46689 | 1.97E-16 | 45 | 101 | IPR009057 | Homeodomain-like |
| TIGRFAMs | TIGR01557 | 9.0E-23 | 46 | 101 | IPR006447 | Myb domain, plants |
| Pfam | PF00249 | 2.4E-9 | 48 | 99 | IPR001005 | SANT/Myb domain |
| Pfam | PF14379 | 1.9E-24 | 147 | 193 | IPR025756 | MYB-CC type transcription factor, LHEQLE-containing domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 420 aa Download sequence Send to blast |
MYHHHQHQGK NIHSSSRMPI PSERHMFLQT GNGSGDSGLV LSTDAKPRLK WTPDLHARFI 60 EAVNQLGGAD KATPKTVMKL MGIPGLTLYH LKSHLQKYRL SKNLHGQSNN VTYKITTSAS 120 TGERLSETNG THMNKLSLGP QANKDLHISE ALQMQIEVQR RLNEQLEVQR HLQLRIEAQG 180 KYLQSVLEKA QETLGRQNLG VVGIEAAKVQ LSELVSKVSS QCLNSAFTEP KDLQGFFPQQ 240 TQTNPPNDCS MDSCLTSSDR SQKEQEIQNG LRHFNSHVFM EHKEATEAPN NLRNPELKWC 300 EDGKKNTFLA PLSKNEERRN YAAESSPNNL SMSIGLERET ENGINLYPER LITESQSDGE 360 FQHRNRIKPE TLKPVDEKVS QDYRLPASYF AAARLDLNTH GDNEAATTCK QLDLNRFSWS |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 6j4k_A | 1e-19 | 45 | 102 | 1 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4k_B | 1e-19 | 45 | 102 | 1 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_A | 1e-19 | 45 | 102 | 1 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_C | 1e-19 | 45 | 102 | 1 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_D | 1e-19 | 45 | 102 | 1 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_F | 1e-19 | 45 | 102 | 1 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_H | 1e-19 | 45 | 102 | 1 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_J | 1e-19 | 45 | 102 | 1 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcriptional activator that may activate the transcription of specific genes involved in nitrogen uptake or assimilation (PubMed:15592750). Acts redundantly with MYR1 as a repressor of flowering and organ elongation under decreased light intensity (PubMed:21255164). Represses gibberellic acid (GA)-dependent responses and affects levels of bioactive GA (PubMed:21255164). {ECO:0000269|PubMed:21255164, ECO:0000305|PubMed:15592750}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00331 | DAP | Transfer from AT3G04030 | Download |
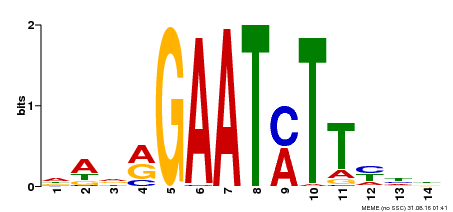
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | KHN27648.1 |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Up-regulated by nitrogen deficiency. {ECO:0000269|PubMed:15592750}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | BT093553 | 0.0 | BT093553.1 Soybean clone JCVI-FLGm-24O19 unknown mRNA. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | NP_001350653.1 | 0.0 | MYB-CC domain-containing transcription factor PHR21 | ||||
| Refseq | XP_028192917.1 | 0.0 | myb-related protein 2-like | ||||
| Swissprot | Q9SQQ9 | 1e-148 | PHL9_ARATH; Myb-related protein 2 | ||||
| TrEMBL | A0A0B2R6R9 | 0.0 | A0A0B2R6R9_GLYSO; Myb family transcription factor APL | ||||
| TrEMBL | I1LTV8 | 0.0 | I1LTV8_SOYBN; Uncharacterized protein | ||||
| STRING | GLYMA12G31020.1 | 0.0 | (Glycine max) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF3852 | 31 | 63 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G04030.3 | 1e-150 | G2-like family protein | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



