 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | KHN36047.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Fabales; Fabaceae; Papilionoideae; Phaseoleae; Glycine; Soja
|
||||||||
| Family | NAC | ||||||||
| Protein Properties | Length: 345aa MW: 39672.4 Da PI: 4.9802 | ||||||||
| Description | NAC family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | NAM | 177.6 | 3.3e-55 | 6 | 134 | 2 | 129 |
NAM 2 ppGfrFhPtdeelvveyLkkkvegkkleleevikevdiykvePwdLpk..kvka.eekewyfFskrdkkyatgkrknratksgyWkatgkdkevlskkg 97
ppGfrFhPtdeelv +yL+kkv+++k++l +vi+e+d+y++ePwdL++ ++ e++ewyfFs++dkky+tg+r+nrat +g+Wkatg+dk+v++ +
KHN36047.1 6 PPGFRFHPTDEELVGYYLRKKVASQKIDL-DVIREIDLYRIEPWDLQErcRIGYdEQNEWYFFSHKDKKYPTGTRTNRATMAGFWKATGRDKAVYE-RA 102
9****************************.9***************953433333667**************************************.99 PP
NAM 98 elvglkktLvfykgrapkgektdWvmheyrle 129
+l+g++ktLvfykgrap+g+k+dW+mheyrle
KHN36047.1 103 KLIGMRKTLVFYKGRAPNGQKSDWIMHEYRLE 134
9*****************************85 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF101941 | 4.84E-59 | 4 | 158 | IPR003441 | NAC domain |
| PROSITE profile | PS51005 | 59.189 | 5 | 158 | IPR003441 | NAC domain |
| Pfam | PF02365 | 1.3E-28 | 6 | 133 | IPR003441 | NAC domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0010200 | Biological Process | response to chitin | ||||
| GO:0048759 | Biological Process | xylem vessel member cell differentiation | ||||
| GO:1901348 | Biological Process | positive regulation of secondary cell wall biogenesis | ||||
| GO:1990110 | Biological Process | callus formation | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0044212 | Molecular Function | transcription regulatory region DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 345 aa Download sequence Send to blast |
MESCIPPGFR FHPTDEELVG YYLRKKVASQ KIDLDVIREI DLYRIEPWDL QERCRIGYDE 60 QNEWYFFSHK DKKYPTGTRT NRATMAGFWK ATGRDKAVYE RAKLIGMRKT LVFYKGRAPN 120 GQKSDWIMHE YRLESDENGP PQASIYEEGW VVCRAFKKKT TGQTKTINIE GWDSSYFYEE 180 ASGASTVVDP IELISRQSQS FVAQNFMCKQ ELEADNLSCM LQDPFVQLPQ LESPSLPLVK 240 RPSTVSLVSD NNEDEDIQNR LLSNNNNTKK VTTDWRALDK FVASQLSQEV DTHESNGVPS 300 SFQPHDDNHD MALLLLQSSR DEGNRLSPFL NTSSDCDIGI CVFEK |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 3ulx_A | 1e-52 | 2 | 159 | 12 | 169 | Stress-induced transcription factor NAC1 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription activator that binds to the secondary wall NAC binding element (SNBE), 5'-(T/A)NN(C/T)(T/C/G)TNNNNNNNA(A/C)GN(A/C/T)(A/T)-3', in the promoter of target genes (By similarity). Involved in xylem formation by promoting the expression of secondary wall-associated transcription factors and of genes involved in secondary wall biosynthesis and programmed cell death, genes driven by the secondary wall NAC binding element (SNBE). Triggers thickening of secondary walls (PubMed:25148240). {ECO:0000250|UniProtKB:Q9LVA1, ECO:0000269|PubMed:25148240}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00266 | DAP | Transfer from AT2G18060 | Download |
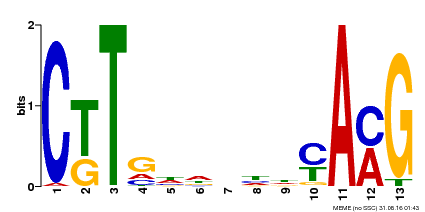
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | KHN36047.1 |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Induced by chitin (e.g. chitooctaose) (PubMed:17722694). Accumulates in plants exposed to callus induction medium (CIM) (PubMed:17581762). {ECO:0000269|PubMed:17581762, ECO:0000269|PubMed:17722694}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | BT094162 | 0.0 | BT094162.1 Soybean clone JCVI-FLGm-18C16 unknown mRNA. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_003517186.1 | 0.0 | NAC domain-containing protein 37 | ||||
| Refseq | XP_006573563.1 | 0.0 | NAC domain-containing protein 37 | ||||
| Refseq | XP_028241587.1 | 0.0 | NAC domain-containing protein 37-like | ||||
| Refseq | XP_028241595.1 | 0.0 | NAC domain-containing protein 37-like | ||||
| Swissprot | Q9SL41 | 1e-157 | NAC37_ARATH; NAC domain-containing protein 37 | ||||
| TrEMBL | A0A445M419 | 0.0 | A0A445M419_GLYSO; NAC domain-containing protein 37 isoform A | ||||
| TrEMBL | K7K496 | 0.0 | K7K496_SOYBN; Uncharacterized protein | ||||
| STRING | GLYMA01G37310.2 | 0.0 | (Glycine max) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF4565 | 34 | 59 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G18060.1 | 1e-148 | vascular related NAC-domain protein 1 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



