 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | KHN36918.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Fabales; Fabaceae; Papilionoideae; Phaseoleae; Glycine; Soja
|
||||||||
| Family | BES1 | ||||||||
| Protein Properties | Length: 331aa MW: 35841 Da PI: 8.3634 | ||||||||
| Description | BES1 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | DUF822 | 212.6 | 9.9e-66 | 2 | 145 | 1 | 150 |
DUF822 1 ggsgrkptwkErEnnkrRERrRRaiaakiyaGLRaqGnyklpkraDnneVlkALcreAGwvvedDGttyrkgskpleeaeaagssasaspesslqsslk 99
++++r pt kErEnnkrRERrRRaiaaki+aGLR++Gn+klpk++DnneVlkALc+eAGw+ve+DGttyrkg+kple+++++g s+ asp+ss++
KHN36918.1 2 TSGTRLPTSKERENNKRRERRRRAIAAKIFAGLRMYGNFKLPKHCDNNEVLKALCNEAGWTVEPDGTTYRKGCKPLERMDIVGGSSAASPCSSYH---- 96
5899*******************************************************************************************.... PP
DUF822 100 ssalaspvesysaspksssfpspssldsislasa..asllpvlsvlslvsssl 150
+sp++sy++sp sssfpspss+ +++ +a +sl+p+l++ls++sss+
KHN36918.1 97 ----PSPCASYNPSPGSSSFPSPSSSPYTQIPNAdgNSLIPWLKNLSTASSSA 145
....*******************9998888776677**********9988765 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF05687 | 2.4E-64 | 4 | 139 | IPR008540 | BES1/BZR1 plant transcription factor, N-terminal |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 331 aa Download sequence Send to blast |
MTSGTRLPTS KERENNKRRE RRRRAIAAKI FAGLRMYGNF KLPKHCDNNE VLKALCNEAG 60 WTVEPDGTTY RKGCKPLERM DIVGGSSAAS PCSSYHPSPC ASYNPSPGSS SFPSPSSSPY 120 TQIPNADGNS LIPWLKNLST ASSSASSPKL PHLYLHSGSI SAPVTPPLSS PTSRTPRINV 180 EWDEQSARPG WTRQQHYSFL PSSSPPSPGR QVVDPEWFAG IKLPHVSPTS PTFSLVSSNP 240 FAFKEDGLPG SGSRMWTPAQ SGTCSPAIPP GSDQNADIPM SEAVSDEFAF GSNMLGLVKP 300 WEGERIHEEF GSDDLELTLG NSKTRYSTII T |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 5zd4_A | 5e-22 | 25 | 77 | 391 | 443 | Maltose-binding periplasmic protein,Protein BRASSINAZOLE-RESISTANT 1 |
| 5zd4_B | 5e-22 | 25 | 77 | 391 | 443 | Maltose-binding periplasmic protein,Protein BRASSINAZOLE-RESISTANT 1 |
| 5zd4_C | 5e-22 | 25 | 77 | 391 | 443 | Maltose-binding periplasmic protein,Protein BRASSINAZOLE-RESISTANT 1 |
| 5zd4_D | 5e-22 | 25 | 77 | 391 | 443 | Maltose-binding periplasmic protein,Protein BRASSINAZOLE-RESISTANT 1 |
| Search in ModeBase | ||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00248 | DAP | Transfer from AT1G78700 | Download |
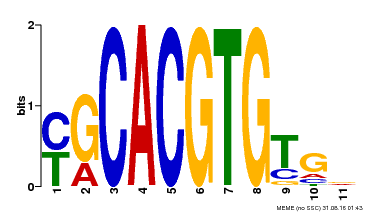
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | KHN36918.1 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AP015040 | 0.0 | AP015040.1 Vigna angularis var. angularis DNA, chromosome 7, almost complete sequence, cultivar: Shumari. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_028191471.1 | 0.0 | BES1/BZR1 homolog protein 4-like | ||||
| Swissprot | Q9ZV88 | 1e-138 | BEH4_ARATH; BES1/BZR1 homolog protein 4 | ||||
| TrEMBL | A0A445IAV6 | 0.0 | A0A445IAV6_GLYSO; BES1/BZR1-like protein 4 isoform A | ||||
| STRING | GLYMA13G34170.1 | 0.0 | (Glycine max) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF2661 | 33 | 75 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G78700.1 | 1e-112 | BES1/BZR1 homolog 4 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



