 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | KHN37282.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Fabales; Fabaceae; Papilionoideae; Phaseoleae; Glycine; Soja
|
||||||||
| Family | C2H2 | ||||||||
| Protein Properties | Length: 325aa MW: 35133.1 Da PI: 10.509 | ||||||||
| Description | C2H2 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | zf-C2H2 | 15.4 | 5.2e-05 | 202 | 225 | 1 | 23 |
EEETTTTEEESSHHHHHHHHHH.T CS
zf-C2H2 1 ykCpdCgksFsrksnLkrHirt.H 23
y+Cp+Cg +F ++ + H ++ H
KHN37282.1 202 YTCPKCGHVFETSQRFAAHVSSsH 225
9******************99888 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS50157 | 9.494 | 202 | 230 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 0.0069 | 202 | 225 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE pattern | PS00028 | 0 | 204 | 225 | IPR007087 | Zinc finger, C2H2 |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0046872 | Molecular Function | metal ion binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 325 aa Download sequence Send to blast |
MALNSPNSSN FSTALSLGSA SNTNEALTVK GMGQLGLNQR ITKVRKSSLT AKERSCLVHG 60 GSENNTESII RASQIVRAQL VHAVSSTPSP VSSNNVVVVV ADKKKAIEIL QAKLLCASAF 120 APKTSPTSLR PISLSHRLTS TPPLKTRRGF LNDDDFAYSL CPSPLSVCFP HETSTLQNHV 180 RGGDTKYDGR MHSYPYKKNG PYTCPKCGHV FETSQRFAAH VSSSHYKYET KSERKKRLMA 240 KIRKRNLRIE WVSGGLTVVP DDAPPTSAFA YSARNNNNVA PPPPVRVVKV EGQSGMELQL 300 APPPGWEKLT AGGGAVKIKL EPVAS |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00523 | DAP | Transfer from AT5G22990 | Download |
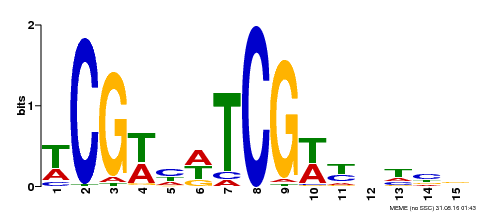
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | KHN37282.1 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AC235297 | 0.0 | AC235297.1 Glycine max strain Williams 82 clone GM_WBb0060P19, complete sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_028183975.1 | 0.0 | uncharacterized protein LOC114370783 | ||||
| TrEMBL | A0A445IUK6 | 0.0 | A0A445IUK6_GLYSO; Uncharacterized protein | ||||
| STRING | GLYMA10G44160.1 | 0.0 | (Glycine max) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF16742 | 7 | 7 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G22990.1 | 4e-23 | C2H2-like zinc finger protein | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



