 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | KHN39136.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Fabales; Fabaceae; Papilionoideae; Phaseoleae; Glycine; Soja
|
||||||||
| Family | WRKY | ||||||||
| Protein Properties | Length: 372aa MW: 40772.7 Da PI: 7.3413 | ||||||||
| Description | WRKY family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | WRKY | 106.2 | 1.7e-33 | 191 | 248 | 2 | 59 |
--SS-EEEEEEE--TT-SS-EEEEEE-STT---EEEEEE-SSSTTEEEEEEES--SS- CS
WRKY 2 dDgynWrKYGqKevkgsefprsYYrCtsagCpvkkkversaedpkvveitYegeHnhe 59
+Dgy+WrKYGqK vk+s++prsYYrCt+++C+vkk+vers +dp++v++tYeg+Hnh+
KHN39136.1 191 EDGYRWRKYGQKAVKNSPYPRSYYRCTTQKCTVKKRVERSFQDPTTVITTYEGQHNHP 248
8********************************************************7 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:2.20.25.80 | 7.1E-35 | 175 | 250 | IPR003657 | WRKY domain |
| SuperFamily | SSF118290 | 2.09E-29 | 182 | 250 | IPR003657 | WRKY domain |
| PROSITE profile | PS50811 | 32.983 | 185 | 250 | IPR003657 | WRKY domain |
| SMART | SM00774 | 7.5E-39 | 190 | 249 | IPR003657 | WRKY domain |
| Pfam | PF03106 | 8.0E-26 | 191 | 248 | IPR003657 | WRKY domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 372 aa Download sequence Send to blast |
MSVEPKELYY QDLVLDHNII NNQHQGTEAG SNTTMYEKHF SSTLIPSVSA AYDSSQAFDP 60 SYMSFTDCLQ GSMDYNSLAT SFGLSPSSSE VFSSVEGNQK PAEGGGDGGG GGGGGSGGSE 120 TLATLNSSIS SSSSEAGAEE DSGKSKKERQ VKTEEGGENS KKGNKEKKKG EKKQKEPRFA 180 FMTKSEVDHL EDGYRWRKYG QKAVKNSPYP RSYYRCTTQK CTVKKRVERS FQDPTTVITT 240 YEGQHNHPVP TSLRGNAAAG MFTPSSLLAT PTHPLAAGSN FPQDLFLHMH YPRHHHQYHH 300 IHNNLFATQS MTNATTATTT TTTATIAPSS FYSSYNNINN SLLNNQFLPP EYGLLQDIVP 360 SMLHNKSHHN QN |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1wj2_A | 1e-28 | 180 | 250 | 7 | 77 | Probable WRKY transcription factor 4 |
| 2lex_A | 1e-28 | 180 | 250 | 7 | 77 | Probable WRKY transcription factor 4 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
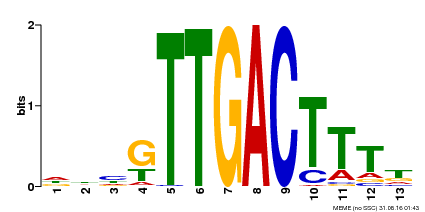
| UniProt | Transcription factor. Interacts specifically with the W box (5'-(T)TGAC[CT]-3'), a frequently occurring elicitor-responsive cis-acting element (By similarity). {ECO:0000250}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00440 | DAP | Transfer from AT4G18170 | Download |
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | KHN39136.1 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AP015034 | 1e-108 | AP015034.1 Vigna angularis var. angularis DNA, chromosome 1, almost complete sequence, cultivar: Shumari. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_028245161.1 | 0.0 | WRKY transcription factor 28-like isoform X1 | ||||
| Swissprot | Q8VWJ2 | 1e-71 | WRK28_ARATH; WRKY transcription factor 28 | ||||
| TrEMBL | A0A0B2RXG9 | 0.0 | A0A0B2RXG9_GLYSO; Putative WRKY transcription factor 28 | ||||
| STRING | GLYMA08G08720.2 | 0.0 | (Glycine max) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF2231 | 34 | 83 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G18170.1 | 4e-50 | WRKY DNA-binding protein 28 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



