 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | KHN39244.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Fabales; Fabaceae; Papilionoideae; Phaseoleae; Glycine; Soja
|
||||||||
| Family | HD-ZIP | ||||||||
| Protein Properties | Length: 838aa MW: 92431.7 Da PI: 6.437 | ||||||||
| Description | HD-ZIP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Homeobox | 57.7 | 2e-18 | 17 | 75 | 3 | 57 |
--SS--HHHHHHHHHHHHHSSS--HHHHHHHHHHC....TS-HHHHHHHHHHHHHHHHC CS
Homeobox 3 kRttftkeqleeLeelFeknrypsaeereeLAkkl....gLterqVkvWFqNrRakekk 57
k ++t+eq+e+Le+l++ +++ps +r++L +++ +++ +q+kvWFqNrR +ek+
KHN39244.1 17 KYVRYTPEQVEALERLYHDCPKPSSIRRQQLIRECpilsNIEPKQIKVWFQNRRCREKQ 75
5679*****************************************************97 PP
| |||||||
| 2 | START | 182.4 | 2.5e-57 | 162 | 370 | 2 | 205 |
HHHHHHHHHHHHHHHC-TT-EEEEEXCCTTEEEEEEESSS.SCEEEEEEEECCSCHHHHHHHHHCCCGGCT-TT-SEEEEEEEECTT..EEEEEEEEXX CS
START 2 laeeaaqelvkkalaeepgWvkssesengdevlqkfeeskvdsgealrasgvvdmvlallveellddkeqWdetlakaetlevissg..galqlmvael 98
+aee+++e+++ka+ ++ Wv+++ +++g++++ +++ s++++g a+ra+g+v +++ v+e+l+d + W ++++++++l+v+ ++ g+++l +++l
KHN39244.1 162 IAEETLAEFLSKATGTAVEWVQMPGMKPGPDSIGIVAISHGCTGVAARACGLVGLEPT-RVAEILKDQPLWFHDCRAVDVLNVLPTAngGTIELLYMQL 259
7899******************************************************.9999999999****************9999********** PP
TTXX-SSX.EEEEEEEEEEE.TTS-EEEEEEEEE-TTS--....-TTSEE-EESSEEEEEEEECTCEEEEEEEE-EE--SSXXHHHHHHHHHHHHHHHH CS
START 99 qalsplvp.RdfvfvRyirqlgagdwvivdvSvdseqkppe...sssvvRaellpSgiliepksnghskvtwvehvdlkgrlphwllrslvksglaega 193
+a+++l+p Rdf+ +Ry+ l++g++vi+++S+ ++q+ p+ +++vRae+lpSg+li+p+++g+s +++v+h+dl+ ++++++lr+l++s+++ ++
KHN39244.1 260 YAPTTLAPaRDFWLLRYTSVLEDGSLVICERSLKNTQNGPSmppVQHFVRAEMLPSGYLIRPCEGGGSIIHIVDHMDLEPWSVPEVLRPLYESSTVLAQ 358
***************************************9988899***************************************************** PP
HHHHHHTXXXXX CS
START 194 ktwvatlqrqce 205
kt++a+l+++++
KHN39244.1 359 KTTMAALRHLRQ 370
*******99876 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS50071 | 15.564 | 12 | 76 | IPR001356 | Homeobox domain |
| SMART | SM00389 | 1.1E-15 | 14 | 80 | IPR001356 | Homeobox domain |
| SuperFamily | SSF46689 | 1.41E-16 | 16 | 78 | IPR009057 | Homeodomain-like |
| CDD | cd00086 | 1.31E-16 | 17 | 77 | No hit | No description |
| Pfam | PF00046 | 5.8E-16 | 18 | 75 | IPR001356 | Homeobox domain |
| Gene3D | G3DSA:1.10.10.60 | 3.5E-18 | 19 | 75 | IPR009057 | Homeodomain-like |
| CDD | cd14686 | 2.37E-6 | 69 | 108 | No hit | No description |
| PROSITE profile | PS50848 | 24.995 | 152 | 366 | IPR002913 | START domain |
| CDD | cd08875 | 1.39E-79 | 156 | 371 | No hit | No description |
| Gene3D | G3DSA:3.30.530.20 | 9.4E-23 | 161 | 367 | IPR023393 | START-like domain |
| SMART | SM00234 | 1.3E-42 | 161 | 371 | IPR002913 | START domain |
| SuperFamily | SSF55961 | 2.75E-38 | 161 | 371 | No hit | No description |
| Pfam | PF01852 | 7.3E-55 | 162 | 370 | IPR002913 | START domain |
| SuperFamily | SSF55961 | 4.62E-5 | 398 | 492 | No hit | No description |
| SuperFamily | SSF55961 | 4.62E-5 | 522 | 597 | No hit | No description |
| Pfam | PF08670 | 3.4E-50 | 695 | 837 | IPR013978 | MEKHLA |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009965 | Biological Process | leaf morphogenesis | ||||
| GO:0010014 | Biological Process | meristem initiation | ||||
| GO:0010075 | Biological Process | regulation of meristem growth | ||||
| GO:0010087 | Biological Process | phloem or xylem histogenesis | ||||
| GO:0048263 | Biological Process | determination of dorsal identity | ||||
| GO:0080060 | Biological Process | integument development | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0008289 | Molecular Function | lipid binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 838 aa Download sequence Send to blast |
MSCKDGSRNG IGMDNGKYVR YTPEQVEALE RLYHDCPKPS SIRRQQLIRE CPILSNIEPK 60 QIKVWFQNRR CREKQRKESS RLQAVNRKLT AMNKLLMEEN DRLQKQVSQL VYENGYFRQH 120 TQITTQATKD TNCESVVTSG QQHNLITQHP PRDASPAGLL SIAEETLAEF LSKATGTAVE 180 WVQMPGMKPG PDSIGIVAIS HGCTGVAARA CGLVGLEPTR VAEILKDQPL WFHDCRAVDV 240 LNVLPTANGG TIELLYMQLY APTTLAPARD FWLLRYTSVL EDGSLVICER SLKNTQNGPS 300 MPPVQHFVRA EMLPSGYLIR PCEGGGSIIH IVDHMDLEPW SVPEVLRPLY ESSTVLAQKT 360 TMAALRHLRQ ISHEVSQSNV TGWGRRPAAL RALSQRLSRG FNEALNGFTD EGWTTIGNDG 420 VDDVTILVNS SPDKLMGLNL SFANGFPSVS NAVLCAKASM LLQNVPPAIL LRFLREHRSE 480 WADNNMDAYT AAAIKVGPCS LSGSRVGNYG GQVILPLAHT IEHEEFLEVI KLEGIAHSPE 540 DTIMPREMFL LQLCSGMDEN AVGTCAELIS APIDASFADD APLLPSGFRI IPLESGKEAS 600 SPNRTLDLAS ALDVGPSGNR ASNGCANSSC MRSVMTIAFE FAFESHMQEH VASMARQYVR 660 SIISSVQRVA LALSPSHLSS HAGLRSPLGT PEAQTLAHWI CNSYRCYLGV ELLKSNNEGN 720 ESLLKSLWHH SDAILCCTLK ALPVFTFSNQ AGLDMLETTL VALQDITLEK IFDDHGRKIL 780 FSEFPQIIQQ GFACLQGGIC LSSMGRPVSY ERVVAWKVLN EEENAHCICF MFVNWSFV |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcription factor involved in the regulation of meristem development to promote lateral organ formation. May regulates procambial and vascular tissue formation or maintenance, and vascular development in inflorescence stems. {ECO:0000269|PubMed:15598805, ECO:0000269|PubMed:15705957, ECO:0000269|PubMed:16617092}. | |||||
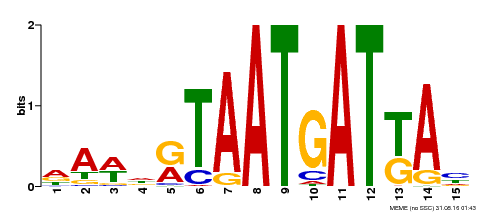
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00200 | DAP | Transfer from AT1G52150 | Download |
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | KHN39244.1 |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: By auxin. Repressed by miR165 and miR166. {ECO:0000269|PubMed:15773855, ECO:0000269|PubMed:16033795, ECO:0000269|PubMed:16617092, ECO:0000269|PubMed:17237362}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_003530109.1 | 0.0 | homeobox-leucine zipper protein ATHB-15 | ||||
| Refseq | XP_028238900.1 | 0.0 | homeobox-leucine zipper protein ATHB-15-like | ||||
| Swissprot | Q9ZU11 | 0.0 | ATB15_ARATH; Homeobox-leucine zipper protein ATHB-15 | ||||
| TrEMBL | A0A445JQZ3 | 0.0 | A0A445JQZ3_GLYSO; Homeobox-leucine zipper protein ATHB-15 isoform D | ||||
| STRING | GLYMA07G01940.1 | 0.0 | (Glycine max) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF1946 | 33 | 87 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G52150.1 | 0.0 | HD-ZIP family protein | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



