 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | KHN41758.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Fabales; Fabaceae; Papilionoideae; Phaseoleae; Glycine; Soja
|
||||||||
| Family | BES1 | ||||||||
| Protein Properties | Length: 664aa MW: 75156.2 Da PI: 6.7465 | ||||||||
| Description | BES1 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | DUF822 | 158.9 | 3.4e-49 | 40 | 184 | 1 | 141 |
DUF822 1 ggsgrkptwkErEnnkrRERrRRaiaakiyaGLRaqGnyklpkraDnneVlkALcreAGwvvedDGttyrkgskpleeaeaagssasaspesslq.ssl 98
gg+g+k+++kE+E++k+RER+RRai+++++aGLR++Gn++lp+raD+n+Vl+AL+reAGwvv++DGttyr+ + p+ + +gs+a +s+es+l+ sl
KHN41758.1 40 GGKGKKEREKEKERTKLRERHRRAITSRMLAGLRQYGNFPLPARADMNDVLAALAREAGWVVDADGTTYRQ-CPPP---SNVGSFAARSVESQLSgGSL 134
689*******************************************************************5.4444...69**************9*** PP
DUF822 99 kssalaspvesysaspksssfpspssldsislasa.......asllpvls 141
++++++ +e++ a+++ ++ +sp+s+ds+ +a+ ++ +p+ +
KHN41758.1 135 RNCSVKETIENQTAVLRIDECLSPASIDSVVIAERdsknekyTNARPINT 184
********************************988666555555555555 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF05687 | 2.8E-50 | 41 | 188 | IPR008540 | BES1/BZR1 plant transcription factor, N-terminal |
| SuperFamily | SSF51445 | 2.85E-155 | 229 | 661 | IPR017853 | Glycoside hydrolase superfamily |
| Gene3D | G3DSA:3.20.20.80 | 9.0E-170 | 231 | 662 | IPR013781 | Glycoside hydrolase, catalytic domain |
| Pfam | PF01373 | 4.6E-87 | 252 | 628 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 8.4E-58 | 269 | 283 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 8.4E-58 | 290 | 308 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 8.4E-58 | 312 | 333 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 8.4E-58 | 405 | 427 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 8.4E-58 | 478 | 497 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 8.4E-58 | 512 | 528 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 8.4E-58 | 529 | 540 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 8.4E-58 | 547 | 570 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 8.4E-58 | 585 | 607 | IPR001554 | Glycoside hydrolase, family 14 |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0000272 | Biological Process | polysaccharide catabolic process | ||||
| GO:0048831 | Biological Process | regulation of shoot system development | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0016161 | Molecular Function | beta-amylase activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 664 aa Download sequence Send to blast |
MALSLVVRNT VSVKCTKREE APPSTRPRGF AAAPVTTNTG GKGKKEREKE KERTKLRERH 60 RRAITSRMLA GLRQYGNFPL PARADMNDVL AALAREAGWV VDADGTTYRQ CPPPSNVGSF 120 AARSVESQLS GGSLRNCSVK ETIENQTAVL RIDECLSPAS IDSVVIAERD SKNEKYTNAR 180 PINTVDCLEA DQFLNVFVLV SAKLDSFGIQ PKSFSLMQDI HSGVHENDFT STPYVSVYVK 240 LPAGIINKFC QLIDPEGIKQ ELIHIKSLNV DGVVVDCWWG IVEGWSSQKY VWSGYRELFN 300 IIREFKLKLQ VVMAFHECGG NDSSDALISL PQWVLDIGKD NQDIFFTDRE GRRNTECLSW 360 GIDKERVLKG RTGIEVYFDM MRSFRTEFDD LFAEGLISAV EVGLGASGEL KYPSFSERMG 420 WRYPGIGEFQ CYDKYLQNSL RRAAKLHGHS FWARGPDNAG HYNSMPHETG FFCERGDYDN 480 YYGRFFLHWY SQTLIDHADN VLSLATLAFE ETKIIVKVPA VYWWYKTPSH AAELTAGYHN 540 PTYQDGYSPV FEVLRKHAVT MKFVCLGFHL SSQEAYEPLI DPEGLSWQVL NSAWDRGLMA 600 AGENALLCYG REGYKRLVEM AKPRNDPDCR HFSFFVYQQP SLLQANVCLS ELDFFVKCMH 660 GKDP |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1wdq_A | 1e-125 | 234 | 624 | 11 | 401 | Beta-amylase |
| Search in ModeBase | ||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00543 | DAP | Transfer from AT5G45300 | Download |
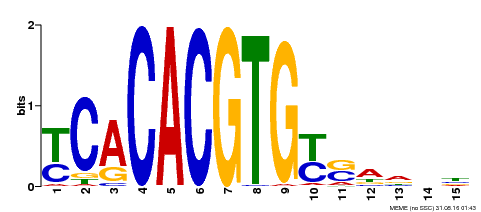
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | KHN41758.1 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_003516502.1 | 0.0 | beta-amylase 8 | ||||
| Refseq | XP_028239751.1 | 0.0 | beta-amylase 8-like | ||||
| Swissprot | Q9FH80 | 0.0 | BAM8_ARATH; Beta-amylase 8 | ||||
| TrEMBL | A0A445M390 | 0.0 | A0A445M390_GLYSO; Beta-amylase | ||||
| TrEMBL | K7K400 | 0.0 | K7K400_SOYBN; Beta-amylase | ||||
| STRING | GLYMA01G35500.2 | 0.0 | (Glycine max) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF11902 | 30 | 32 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G45300.1 | 0.0 | beta-amylase 2 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



