 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | KHN47180.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Fabales; Fabaceae; Papilionoideae; Phaseoleae; Glycine; Soja
|
||||||||
| Family | HD-ZIP | ||||||||
| Protein Properties | Length: 285aa MW: 32672.2 Da PI: 4.4616 | ||||||||
| Description | HD-ZIP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Homeobox | 63.3 | 3.5e-20 | 65 | 119 | 2 | 56 |
T--SS--HHHHHHHHHHHHHSSS--HHHHHHHHHHCTS-HHHHHHHHHHHHHHHH CS
Homeobox 2 rkRttftkeqleeLeelFeknrypsaeereeLAkklgLterqVkvWFqNrRakek 56
+k++++++eq++ Le+ Fe+++++ e++++LAkklgL+ rqV vWFqNrRa++k
KHN47180.1 65 KKKHRLSSEQVHLLEKNFEEENKLEPERKTQLAKKLGLQPRQVAVWFQNRRARWK 119
78999*************************************************9 PP
| |||||||
| 2 | HD-ZIP_I/II | 132.6 | 1.5e-42 | 65 | 156 | 1 | 92 |
HD-ZIP_I/II 1 ekkrrlskeqvklLEesFeeeekLeperKvelareLglqprqvavWFqnrRARtktkqlEkdyeaLkraydalkeenerLekeveeLreelk 92
+kk+rls+eqv+lLE++Feee+kLeperK++la++LglqprqvavWFqnrRAR+ktkqlE+dy++Lk++yd+l ++ +++ ke+e+L++e+
KHN47180.1 65 KKKHRLSSEQVHLLEKNFEEENKLEPERKTQLAKKLGLQPRQVAVWFQNRRARWKTKQLERDYDVLKSSYDTLLSSYDSIMKENEKLKSEVV 156
59**************************************************************************************9986 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF46689 | 1.92E-20 | 49 | 123 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS50071 | 18.641 | 61 | 121 | IPR001356 | Homeobox domain |
| SMART | SM00389 | 6.0E-19 | 63 | 125 | IPR001356 | Homeobox domain |
| Pfam | PF00046 | 1.9E-17 | 65 | 119 | IPR001356 | Homeobox domain |
| CDD | cd00086 | 8.92E-19 | 65 | 122 | No hit | No description |
| Gene3D | G3DSA:1.10.10.60 | 4.0E-22 | 66 | 128 | IPR009057 | Homeodomain-like |
| PRINTS | PR00031 | 4.3E-6 | 92 | 101 | IPR000047 | Helix-turn-helix motif |
| PROSITE pattern | PS00027 | 0 | 96 | 119 | IPR017970 | Homeobox, conserved site |
| PRINTS | PR00031 | 4.3E-6 | 101 | 117 | IPR000047 | Helix-turn-helix motif |
| Pfam | PF02183 | 1.8E-18 | 121 | 163 | IPR003106 | Leucine zipper, homeobox-associated |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0001558 | Biological Process | regulation of cell growth | ||||
| GO:0009637 | Biological Process | response to blue light | ||||
| GO:0009651 | Biological Process | response to salt stress | ||||
| GO:0009965 | Biological Process | leaf morphogenesis | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0042803 | Molecular Function | protein homodimerization activity | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 285 aa Download sequence Send to blast |
MESGRIFFDA SASRGNNMLF LGNTELAFRG RSIMSMEEAS KRRPFFTSPD ELYDEEYYEK 60 QSPEKKKHRL SSEQVHLLEK NFEEENKLEP ERKTQLAKKL GLQPRQVAVW FQNRRARWKT 120 KQLERDYDVL KSSYDTLLSS YDSIMKENEK LKSEVVSLNE KLQVQAKEVP EETLCDKKVD 180 PLPVDEDMAP IFSTRVEDHL SSGSVGSAVV DEGSPQVVVD SVDSYFPADN YGGCMGPVER 240 VQSEQEDGSD DGRSYLDVFV ASETENQNHE EGEALGWWTN MYYVG |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 113 | 121 | RRARWKTKQ |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
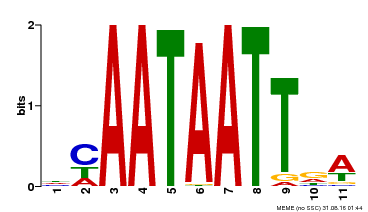
| UniProt | Probable transcription activator involved in leaf development. Binds to the DNA sequence 5'-CAAT[AT]ATTG-3'. {ECO:0000269|PubMed:8535134}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00323 | DAP | Transfer from AT3G01470 | Download |
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | KHN47180.1 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | - | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | BT093728 | 0.0 | BT093728.1 Soybean clone JCVI-FLGm-24I10 unknown mRNA. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_028189510.1 | 0.0 | homeobox-leucine zipper protein HAT5-like isoform X2 | ||||
| Swissprot | Q02283 | 2e-95 | HAT5_ARATH; Homeobox-leucine zipper protein HAT5 | ||||
| TrEMBL | A0A445I616 | 0.0 | A0A445I616_GLYSO; Homeobox-leucine zipper protein HAT5 | ||||
| STRING | GLYMA13G23890.3 | 0.0 | (Glycine max) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF11283 | 30 | 39 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G01470.1 | 8e-77 | homeobox 1 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



