 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | KHN48637.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Fabales; Fabaceae; Papilionoideae; Phaseoleae; Glycine; Soja
|
||||||||
| Family | MYB_related | ||||||||
| Protein Properties | Length: 288aa MW: 31371.9 Da PI: 9.0861 | ||||||||
| Description | MYB_related family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 44.5 | 3.5e-14 | 82 | 126 | 3 | 47 |
SS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHH CS
Myb_DNA-binding 3 rWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqky 47
+WT+eE++l++ + ++ G+g+W+ I++ k+Rt+ q+ s+ qky
KHN48637.1 82 PWTEEEHKLFLVGLQKVGKGDWRGISKNYVKTRTPTQVASHAQKY 126
8*******************************************9 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51294 | 18.262 | 75 | 131 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 2.99E-17 | 77 | 132 | IPR009057 | Homeodomain-like |
| TIGRFAMs | TIGR01557 | 9.1E-17 | 78 | 129 | IPR006447 | Myb domain, plants |
| SMART | SM00717 | 3.9E-10 | 79 | 129 | IPR001005 | SANT/Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 5.9E-11 | 81 | 126 | IPR009057 | Homeodomain-like |
| Pfam | PF00249 | 5.7E-12 | 82 | 126 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 7.58E-10 | 82 | 127 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009651 | Biological Process | response to salt stress | ||||
| GO:0009723 | Biological Process | response to ethylene | ||||
| GO:0009733 | Biological Process | response to auxin | ||||
| GO:0009737 | Biological Process | response to abscisic acid | ||||
| GO:0009739 | Biological Process | response to gibberellin | ||||
| GO:0009751 | Biological Process | response to salicylic acid | ||||
| GO:0009753 | Biological Process | response to jasmonic acid | ||||
| GO:0046686 | Biological Process | response to cadmium ion | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 288 aa Download sequence Send to blast |
MSRASSAADS AASGEIILFG VRVVVDSMRK SVSMNNLSQY EHPLDATTTN NNKDAAGYAS 60 ADDAAPQNSG RLRERERKRG VPWTEEEHKL FLVGLQKVGK GDWRGISKNY VKTRTPTQVA 120 SHAQKYFLRR SNLNRRRRRS SLFDITTDTV SAIPMEEEQV QNQDTLCHSQ QQPVFPAETS 180 KINGFPAMPV YQFGVGSSGV ISVQVQGTGN PMEELTLGQG NVEKHNVPNK VSTVSDIITP 240 SSSSSAVDPP TLSLGLSFSS DQRQTSSRHS ALHAMQCFSN GDSIISVA |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Binds selectively to the DNA sequence 5'-[GA]GATAA-3' and may act as a transcription factor involved in the regulation of drought-responsive genes. Enhances stomatal closure in response to abscisic acid (ABA). Confers drought and salt tolerance. {ECO:0000269|PubMed:21030505}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00631 | PBM | Transfer from PK23829.1 | Download |
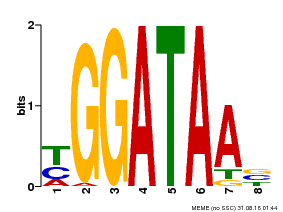
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | KHN48637.1 |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Up-regulated by drought, high salinity, and ABA. {ECO:0000269|PubMed:21030505}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | - | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | FJ550209 | 0.0 | FJ550209.1 Glycine max cultivar Harosoy63 isoflavonoid regulator (MYB176) mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | NP_001236048.2 | 0.0 | transcription factor MYB176 | ||||
| Refseq | XP_028231455.1 | 0.0 | transcription factor MYB1R1-like | ||||
| Swissprot | Q2V9B0 | 4e-68 | MY1R1_SOLTU; Transcription factor MYB1R1 | ||||
| TrEMBL | A0A445KIN1 | 0.0 | A0A445KIN1_GLYSO; Transcription factor MYB1R1 | ||||
| TrEMBL | D8L1Z5 | 0.0 | D8L1Z5_SOYBN; Isoflavonoid regulator | ||||
| STRING | GLYMA05G01640.1 | 0.0 | (Glycine max) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF4248 | 31 | 54 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G74840.2 | 4e-48 | MYB_related family protein | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



