 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | KN538735.1_FGP007 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Oryzoideae; Oryzeae; Oryzinae; Oryza
|
||||||||
| Family | C3H | ||||||||
| Protein Properties | Length: 259aa MW: 28308.2 Da PI: 10.2645 | ||||||||
| Description | C3H family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | zf-CCCH | 48.7 | 1.2e-15 | 185 | 210 | 1 | 26 |
--S---SGGGGTS--TTTTT-SS-SS CS
zf-CCCH 1 yktelCrffartGtCkyGdrCkFaHg 26
+ktelC+ + +tG C+yGd+C+FaHg
KN538735.1_FGP007 185 FKTELCNKWEETGDCPYGDQCQFAHG 210
79***********************9 PP
| |||||||
| 2 | zf-CCCH | 19.3 | 2e-06 | 223 | 248 | 1 | 26 |
--S---SGGGGTS--TTTTT-SS-SS CS
zf-CCCH 1 yktelCrffartGtCkyGdrCkFaHg 26
ykt+ Cr +++ C+yG rC+F H+
KN538735.1_FGP007 223 YKTAVCRMVLAGDVCPYGHRCHFRHS 248
9**********55**********998 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:4.10.1000.10 | 7.7E-19 | 179 | 217 | IPR000571 | Zinc finger, CCCH-type |
| SuperFamily | SSF90229 | 1.44E-10 | 181 | 216 | IPR000571 | Zinc finger, CCCH-type |
| SMART | SM00356 | 2.5E-9 | 184 | 211 | IPR000571 | Zinc finger, CCCH-type |
| PROSITE profile | PS50103 | 15.413 | 184 | 212 | IPR000571 | Zinc finger, CCCH-type |
| Pfam | PF00642 | 1.4E-12 | 185 | 210 | IPR000571 | Zinc finger, CCCH-type |
| SuperFamily | SSF90229 | 3.27E-8 | 218 | 250 | IPR000571 | Zinc finger, CCCH-type |
| Gene3D | G3DSA:4.10.1000.10 | 3.7E-11 | 218 | 248 | IPR000571 | Zinc finger, CCCH-type |
| SMART | SM00356 | 9.5E-5 | 222 | 249 | IPR000571 | Zinc finger, CCCH-type |
| PROSITE profile | PS50103 | 13.009 | 222 | 250 | IPR000571 | Zinc finger, CCCH-type |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0051511 | Biological Process | negative regulation of unidimensional cell growth | ||||
| GO:1901347 | Biological Process | negative regulation of secondary cell wall biogenesis | ||||
| GO:0003729 | Molecular Function | mRNA binding | ||||
| GO:0044212 | Molecular Function | transcription regulatory region DNA binding | ||||
| GO:0046872 | Molecular Function | metal ion binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 259 aa Download sequence Send to blast |
MATGEHKSIL PASPNGKSTH LKVHPVRLAV AAAGRKETHL AHAPTYAAAL RRENAELRVT 60 NSDLARRLAL LSGKHTAAVA VADEIRRLRL GEQKVAADTK ERTPEKLAVL PKSISVRSTS 120 YLKLNQQSQA ATATPAAPNR KPRTSSNPTN PPSSRAYDGG KKGDEQKAQP ADSGAELEVY 180 NQGMFKTELC NKWEETGDCP YGDQCQFAHG VAELRPVIRH PRYKTAVCRM VLAGDVCPYG 240 HRCHFRHSLT PAERLLLRS |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1rgo_A | 2e-21 | 185 | 248 | 4 | 67 | Butyrate response factor 2 |
| Search in ModeBase | ||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00383 | ampDAP | Transfer from AT1G68200 | Download |
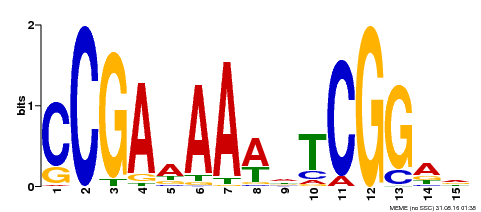
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | KN538735.1_FGP007 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AK108658 | 0.0 | AK108658.1 Oryza sativa Japonica Group cDNA clone:002-148-E11, full insert sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_015631647.1 | 1e-137 | zinc finger CCCH domain-containing protein 9 isoform X2 | ||||
| Swissprot | A2ZVY5 | 1e-136 | C3H9_ORYSJ; Zinc finger CCCH domain-containing protein 9 | ||||
| TrEMBL | A0A0E0C681 | 1e-137 | A0A0E0C681_9ORYZ; Uncharacterized protein | ||||
| TrEMBL | A0A0E0JLQ7 | 1e-137 | A0A0E0JLQ7_ORYPU; Uncharacterized protein | ||||
| STRING | OMERI01G23770.1 | 1e-137 | (Oryza meridionalis) | ||||
| STRING | OPUNC01G24490.1 | 1e-137 | (Oryza punctata) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP2775 | 37 | 84 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G66810.1 | 2e-49 | C3H family protein | ||||



