 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | KN538753.1_FGP048 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Oryzoideae; Oryzeae; Oryzinae; Oryza
|
||||||||
| Family | SBP | ||||||||
| Protein Properties | Length: 638aa MW: 71379.3 Da PI: 6.609 | ||||||||
| Description | SBP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | SBP | 111.9 | 3.9e-35 | 3 | 77 | 2 | 76 |
-SSTT-----TT--HHHHHTT--HHHHT-S-EEETTEEEEE-TTTSSEEETTT--SS--S-STTTT-------S- CS
SBP 2 CqvegCeadlseakeyhrrhkvCevhskapvvlvsgleqrfCqqCsrfhelsefDeekrsCrrrLakhnerrrkk 76
Cqv+gCead+ e+k yhrrh+vC +++a++v+++g+++r+CqqC++fh l +fDe+krsCrr+L++hn+rrr+k
KN538753.1_FGP048 3 CQVPGCEADIRELKGYHRRHRVCLRCAHAAAVMLDGVQKRYCQQCGKFHILLDFDEDKRSCRRKLERHNKRRRRK 77
************************************************************************997 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51141 | 29.521 | 1 | 77 | IPR004333 | Transcription factor, SBP-box |
| Gene3D | G3DSA:4.10.1100.10 | 8.3E-26 | 2 | 64 | IPR004333 | Transcription factor, SBP-box |
| SuperFamily | SSF103612 | 2.35E-33 | 2 | 80 | IPR004333 | Transcription factor, SBP-box |
| Pfam | PF03110 | 4.7E-27 | 3 | 77 | IPR004333 | Transcription factor, SBP-box |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0055070 | Biological Process | copper ion homeostasis | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 638 aa Download sequence Send to blast |
MKCQVPGCEA DIRELKGYHR RHRVCLRCAH AAAVMLDGVQ KRYCQQCGKF HILLDFDEDK 60 RSCRRKLERH NKRRRRKPDS KGILEKDVDD QLDFSADGSG DGELREENID VTTSETLETV 120 LSNKVLDRET PVGSDDVLSS PTCAQPSLQI DQSKSLVTFA ASVEACLGTK QENTKLTNSP 180 VHDTKSTYSS SCPTGRVSFK LYDWNPAEFP RRLRHQIFEW LSSMPVELEG YIRPGCTILT 240 VFVAMPQHMW DKLSEDTGNL VKSLVNAPNS LLLGKGAFFI HVNNMIFQVL KDGATLTSTR 300 LEVQSPRIHY VHPSWFEAGK PIDLILCGSS LDQPKFRSLV SFDGLYLKHD CRRILSHETF 360 DYIGSGEHIL DSQHEIFRIN ITTSKLDTHG PAFVEVENMF GLSNFVPILV GSKHLCSELE 420 QIHDALCGSS DISSDPCELR GLRQTAMSGF LIDIGWLIRK PSIDEFQNLL SLANIQRWIC 480 MMKFLIQNDF INVLEIIVNS LDNIIGSELL SNLEKGRLEN HVTEFLGYRQ VDTRWAGDYA 540 PNQPKLGISV PLAESTGTSG EHDLHSTNEA SGEEENMPLV TKALPHRQCC HPEISARWLN 600 AASIGAFPGG AMRMRLATTV VIGAVSRGAC RASKEVLV |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1ul5_A | 6e-41 | 2 | 82 | 5 | 85 | squamosa promoter binding protein-like 7 |
| Search in ModeBase | ||||||
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 59 | 77 | KRSCRRKLERHNKRRRRKP |
| 2 | 63 | 75 | RRKLERHNKRRRR |
| 3 | 70 | 77 | NKRRRRKP |
| 4 | 71 | 76 | KRRRRK |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
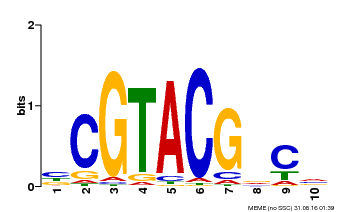
| UniProt | Trans-acting factor that binds specifically to the consensus nucleotide sequence 5'-TNCGTACAA-3'. {ECO:0000250}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00604 | PBM | Transfer from AT5G18830 | Download |
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | KN538753.1_FGP048 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AK100057 | 0.0 | AK100057.1 Oryza sativa Japonica Group cDNA clone:J013158B16, full insert sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_015640052.1 | 0.0 | squamosa promoter-binding-like protein 9 | ||||
| Swissprot | Q6I576 | 0.0 | SPL9_ORYSJ; Squamosa promoter-binding-like protein 9 | ||||
| TrEMBL | B9FPK1 | 0.0 | B9FPK1_ORYSJ; Uncharacterized protein | ||||
| STRING | OGLUM05G17090.1 | 0.0 | (Oryza glumipatula) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP6922 | 33 | 44 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G18830.2 | 1e-137 | squamosa promoter binding protein-like 7 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



