 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | KN538869.1_FGP001 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Oryzoideae; Oryzeae; Oryzinae; Oryza
|
||||||||
| Family | SBP | ||||||||
| Protein Properties | Length: 932aa MW: 101392 Da PI: 5.4696 | ||||||||
| Description | SBP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | SBP | 130.1 | 8e-41 | 102 | 179 | 1 | 78 |
--SSTT-----TT--HHHHHTT--HHHHT-S-EEETTEEEEE-TTTSSEEETTT--SS--S-STTTT-------S--- CS
SBP 1 lCqvegCeadlseakeyhrrhkvCevhskapvvlvsgleqrfCqqCsrfhelsefDeekrsCrrrLakhnerrrkkqa 78
+CqvegC+adl+ +++yhrrhkvCe+h+ka++++v ++ qrfCqqCsrfh l+efDe+krsCrrrLa+hn+rrrk+++
KN538869.1_FGP001 102 ACQVEGCTADLTGVRDYHRRHKVCEMHAKATTAVVGNTVQRFCQQCSRFHPLQEFDEGKRSCRRRLAGHNRRRRKTRP 179
5**************************************************************************875 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:4.10.1100.10 | 6.1E-34 | 96 | 164 | IPR004333 | Transcription factor, SBP-box |
| PROSITE profile | PS51141 | 32.394 | 100 | 177 | IPR004333 | Transcription factor, SBP-box |
| SuperFamily | SSF103612 | 1.7E-38 | 101 | 181 | IPR004333 | Transcription factor, SBP-box |
| Pfam | PF03110 | 1.6E-30 | 103 | 176 | IPR004333 | Transcription factor, SBP-box |
| SuperFamily | SSF48403 | 2.05E-8 | 713 | 819 | IPR020683 | Ankyrin repeat-containing domain |
| Gene3D | G3DSA:1.25.40.20 | 2.4E-7 | 717 | 822 | IPR020683 | Ankyrin repeat-containing domain |
| CDD | cd00204 | 1.95E-8 | 719 | 819 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 932 aa Download sequence Send to blast |
MEAARVGAQS RHFSPSSSEE AGAASVRNVN VRGDSDKRKR VVVIDDDDVE DDELVENGGG 60 SLSLRIGGDA VAHGAGVGGG ADEEDRNGKK IRVQGGSPSG PACQVEGCTA DLTGVRDYHR 120 RHKVCEMHAK ATTAVVGNTV QRFCQQCSRF HPLQEFDEGK RSCRRRLAGH NRRRRKTRPE 180 VAVGGSACTE DKVSSYLLLG LLGVCANLNG NVFMDSTIVL AADNAEHLRG QELLSGLLRN 240 LGAVAKSLDP KELCKLLEAC QSMQDGSNAG TSETANALVN TAVAEAAGPS NSKMPFVNGD 300 QCGLASSSVV PVQSKSPTVA TPDPPACKLK DFDLNDTCGG MEGFEDGYEG SPTPAFKTTD 360 SPNCPSWMHQ DSTQSPPQTS GNSDSTSAQS LSSSNGDAQC RTDKIVFKLF EKVPSDLPPV 420 LRSQILGWLS SSPTDIESYI RPGCIILTVY LRLVESAWKE LSDNMSSNLD KLLNSSTGNF 480 WASGLVFVMV RHQIAFMHNG QLMLDRPLAN SAHHYCKILC VRPIAAPFST KVNFRVEGLN 540 LVSDSSRLIC SFEGSCIFQE DTDNIVDDVE HDDIEYLNFC CPLPSSRGRG FVEVEDDGFS 600 NGFFPFIIAE QDICSEVCEL ESIFESSSHE QADDDNARNQ ALEFLNELGW LLHRANIISK 660 QDKVPLASFN IWRFRNLGIF AMEREWCAVT KLLIDFLFIG LVDIGSKSPE EVVLSENLLH 720 AAVRMKSAQM VRFLLGYKPN ESLKGTAETF LFRPDVQGPS KFTPLHIAAA TDDAEDVLDA 780 LTNDPGLVGI NTWRNARDNA GFTPEDYARQ RGNDAYLNMV EKKINKHLGK GHVVLGVPSS 840 IHPVITDGVK PGEVSLEIGM TVPPPAARCN ACSRQARMYP NSTARTFLYR PAMLTVMGIA 900 VICVCVGLLL HTCPKVYAAP TFRWELLERG PM |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1ul4_A | 1e-30 | 96 | 176 | 4 | 84 | squamosa promoter binding protein-like 4 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
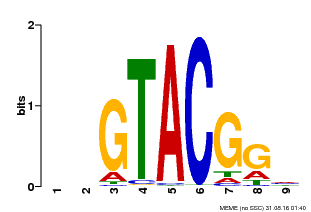
| UniProt | Trans-acting factor that binds specifically to the consensus nucleotide sequence 5'-TNCGTACAA-3'. {ECO:0000250}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00633 | PBM | Transfer from PK06791.1 | Download |
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | KN538869.1_FGP001 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AK072164 | 0.0 | AK072164.1 Oryza sativa Japonica Group cDNA clone:J013130O11, full insert sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_015631511.1 | 0.0 | squamosa promoter-binding-like protein 6 isoform X1 | ||||
| Swissprot | Q75LH6 | 0.0 | SPL6_ORYSJ; Squamosa promoter-binding-like protein 6 | ||||
| TrEMBL | A0A0D9ZFD6 | 0.0 | A0A0D9ZFD6_9ORYZ; Uncharacterized protein | ||||
| STRING | OGLUM03G39810.1 | 0.0 | (Oryza glumipatula) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP1998 | 38 | 92 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G47070.1 | 0.0 | squamosa promoter binding protein-like 1 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



